Tutorial
Introduction to working with NEON eddy flux data
Authors: [Claire K. Lunch]
Last Updated: Mar 12, 2021
This data tutorial provides an introduction to working with NEON eddy
flux data, using the neonUtilities R package. If you are new to NEON
data, we recommend starting with a more general tutorial, such as the
neonUtilities tutorial
or the Download and Explore tutorial.
Some of the functions and techniques described in those tutorials will
be used here, as well as functions and data formats that are unique to
the eddy flux system.
This tutorial assumes general familiarity with eddy flux data and associated concepts.
1. Setup
Start by installing and loading packages and setting options.
To work with the NEON flux data, we need the rhdf5 package,
which is hosted on Bioconductor, and requires a different
installation process than CRAN packages:
install.packages('BiocManager')
BiocManager::install('rhdf5')
install.packages('neonUtilities')
options(stringsAsFactors=F)
library(neonUtilities)
Use the zipsByProduct() function from the neonUtilities package to
download flux data from two sites and two months. The transformations
and functions below will work on any time range and site(s), but two
sites and two months allows us to see all the available functionality
while minimizing download size.
Inputs to the zipsByProduct() function:
-
dpID: DP4.00200.001, the bundled eddy covariance product -
package: basic (the expanded package is not covered in this tutorial) -
site: NIWO = Niwot Ridge and HARV = Harvard Forest -
startdate: 2018-06 (both dates are inclusive) -
enddate: 2018-07 (both dates are inclusive) -
savepath: modify this to something logical on your machine -
check.size: T if you want to see file size before downloading, otherwise F
The download may take a while, especially if you're on a slow network. For faster downloads, consider using an API token.
zipsByProduct(dpID="DP4.00200.001", package="basic",
site=c("NIWO", "HARV"),
startdate="2018-06", enddate="2018-07",
savepath="~/Downloads",
check.size=F)
2. Data Levels
There are five levels of data contained in the eddy flux bundle. For full details, refer to the NEON algorithm document.
Briefly, the data levels are:
- Level 0' (dp0p): Calibrated raw observations
- Level 1 (dp01): Time-aggregated observations, e.g. 30-minute mean gas concentrations
- Level 2 (dp02): Time-interpolated data, e.g. rate of change of a gas concentration
- Level 3 (dp03): Spatially interpolated data, i.e. vertical profiles
- Level 4 (dp04): Fluxes
The dp0p data are available in the expanded data package and are beyond the scope of this tutorial.
The dp02 and dp03 data are used in storage calculations, and the dp04 data include both the storage and turbulent components. Since many users will want to focus on the net flux data, we'll start there.
3. Extract Level 4 data (Fluxes!)
To extract the Level 4 data from the HDF5 files and merge them into a
single table, we'll use the stackEddy() function from the neonUtilities
package.
stackEddy() requires two inputs:
-
filepath: Path to a file or folder, which can be any one of:- A zip file of eddy flux data downloaded from the NEON data portal
- A folder of eddy flux data downloaded by the
zipsByProduct()function - The folder of files resulting from unzipping either of 1 or 2
- One or more HDF5 files of NEON eddy flux data
-
level: dp01-4
Input the filepath you downloaded to using zipsByProduct() earlier,
including the filestoStack00200 folder created by the function, and
dp04:
flux <- stackEddy(filepath="~/Downloads/filesToStack00200",
level="dp04")
We now have an object called flux. It's a named list containing four
tables: one table for each site's data, and variables and objDesc
tables.
names(flux)
## [1] "HARV" "NIWO" "variables" "objDesc"
Let's look at the contents of one of the site data files:
head(flux$NIWO)
## timeBgn timeEnd data.fluxCo2.nsae.flux data.fluxCo2.stor.flux data.fluxCo2.turb.flux
## 1 2018-06-01 00:00:00 2018-06-01 00:29:59 0.1713858 -0.06348163 0.2348674
## 2 2018-06-01 00:30:00 2018-06-01 00:59:59 0.9251711 0.08748146 0.8376896
## 3 2018-06-01 01:00:00 2018-06-01 01:29:59 0.5005812 0.02231698 0.4782642
## 4 2018-06-01 01:30:00 2018-06-01 01:59:59 0.8032820 0.25569306 0.5475889
## 5 2018-06-01 02:00:00 2018-06-01 02:29:59 0.4897685 0.23090472 0.2588638
## 6 2018-06-01 02:30:00 2018-06-01 02:59:59 0.9223979 0.06228581 0.8601121
## data.fluxH2o.nsae.flux data.fluxH2o.stor.flux data.fluxH2o.turb.flux data.fluxMome.turb.veloFric
## 1 15.876622 3.3334970 12.543125 0.2047081
## 2 8.089274 -1.2063258 9.295600 0.1923735
## 3 5.290594 -4.4190781 9.709672 0.1200918
## 4 9.190214 0.2030371 8.987177 0.1177545
## 5 3.111909 0.1349363 2.976973 0.1589189
## 6 4.613676 -0.3929445 5.006621 0.1114406
## data.fluxTemp.nsae.flux data.fluxTemp.stor.flux data.fluxTemp.turb.flux data.foot.stat.angZaxsErth
## 1 4.7565505 -1.4575094 6.2140599 94.2262
## 2 -0.2717454 0.3403877 -0.6121331 355.4252
## 3 -4.2055147 0.1870677 -4.3925824 359.8013
## 4 -13.3834484 -2.4904300 -10.8930185 137.7743
## 5 -5.1854815 -0.7514531 -4.4340284 188.4799
## 6 -7.7365481 -1.9046775 -5.8318707 183.1920
## data.foot.stat.distReso data.foot.stat.veloYaxsHorSd data.foot.stat.veloZaxsHorSd data.foot.stat.veloFric
## 1 8.34 0.7955893 0.2713232 0.2025427
## 2 8.34 0.8590177 0.2300000 0.2000000
## 3 8.34 1.2601763 0.2300000 0.2000000
## 4 8.34 0.7332641 0.2300000 0.2000000
## 5 8.34 0.7096286 0.2300000 0.2000000
## 6 8.34 0.3789859 0.2300000 0.2000000
## data.foot.stat.distZaxsMeasDisp data.foot.stat.distZaxsRgh data.foot.stat.distZaxsAbl
## 1 8.34 0.04105708 1000
## 2 8.34 0.27991938 1000
## 3 8.34 0.21293225 1000
## 4 8.34 0.83400000 1000
## 5 8.34 0.83400000 1000
## 6 8.34 0.83400000 1000
## data.foot.stat.distXaxs90 data.foot.stat.distXaxsMax data.foot.stat.distYaxs90 qfqm.fluxCo2.nsae.qfFinl
## 1 325.26 133.44 25.02 1
## 2 266.88 108.42 50.04 1
## 3 275.22 116.76 66.72 1
## 4 208.50 83.40 75.06 1
## 5 208.50 83.40 66.72 1
## 6 208.50 83.40 41.70 1
## qfqm.fluxCo2.stor.qfFinl qfqm.fluxCo2.turb.qfFinl qfqm.fluxH2o.nsae.qfFinl qfqm.fluxH2o.stor.qfFinl
## 1 1 1 1 1
## 2 1 1 1 0
## 3 1 1 1 0
## 4 1 1 1 0
## 5 1 1 1 0
## 6 1 1 1 1
## qfqm.fluxH2o.turb.qfFinl qfqm.fluxMome.turb.qfFinl qfqm.fluxTemp.nsae.qfFinl qfqm.fluxTemp.stor.qfFinl
## 1 1 0 0 0
## 2 1 0 1 0
## 3 1 1 0 0
## 4 1 1 0 0
## 5 1 0 0 0
## 6 1 0 0 0
## qfqm.fluxTemp.turb.qfFinl qfqm.foot.turb.qfFinl
## 1 0 0
## 2 1 0
## 3 0 0
## 4 0 0
## 5 0 0
## 6 0 0
The variables and objDesc tables can help you interpret the column
headers in the data table. The objDesc table contains definitions for
many of the terms used in the eddy flux data product, but it isn't
complete. To get the terms of interest, we'll break up the column headers
into individual terms and look for them in the objDesc table:
term <- unlist(strsplit(names(flux$NIWO), split=".", fixed=T))
flux$objDesc[which(flux$objDesc$Object %in% term),]
## Object
## 138 angZaxsErth
## 171 data
## 343 qfFinl
## 420 qfqm
## 604 timeBgn
## 605 timeEnd
## Description
## 138 Wind direction
## 171 Represents data fields
## 343 The final quality flag indicating if the data are valid for the given aggregation period (1=fail, 0=pass)
## 420 Quality flag and quality metrics, represents quality flags and quality metrics that accompany the provided data
## 604 The beginning time of the aggregation period
## 605 The end time of the aggregation period
For the terms that aren't captured here, fluxCo2, fluxH2o, and fluxTemp
are self-explanatory. The flux components are
-
turb: Turbulent flux -
stor: Storage -
nsae: Net surface-atmosphere exchange
The variables table contains the units for each field:
flux$variables
## category system variable stat units
## 1 data fluxCo2 nsae timeBgn NA
## 2 data fluxCo2 nsae timeEnd NA
## 3 data fluxCo2 nsae flux umolCo2 m-2 s-1
## 4 data fluxCo2 stor timeBgn NA
## 5 data fluxCo2 stor timeEnd NA
## 6 data fluxCo2 stor flux umolCo2 m-2 s-1
## 7 data fluxCo2 turb timeBgn NA
## 8 data fluxCo2 turb timeEnd NA
## 9 data fluxCo2 turb flux umolCo2 m-2 s-1
## 10 data fluxH2o nsae timeBgn NA
## 11 data fluxH2o nsae timeEnd NA
## 12 data fluxH2o nsae flux W m-2
## 13 data fluxH2o stor timeBgn NA
## 14 data fluxH2o stor timeEnd NA
## 15 data fluxH2o stor flux W m-2
## 16 data fluxH2o turb timeBgn NA
## 17 data fluxH2o turb timeEnd NA
## 18 data fluxH2o turb flux W m-2
## 19 data fluxMome turb timeBgn NA
## 20 data fluxMome turb timeEnd NA
## 21 data fluxMome turb veloFric m s-1
## 22 data fluxTemp nsae timeBgn NA
## 23 data fluxTemp nsae timeEnd NA
## 24 data fluxTemp nsae flux W m-2
## 25 data fluxTemp stor timeBgn NA
## 26 data fluxTemp stor timeEnd NA
## 27 data fluxTemp stor flux W m-2
## 28 data fluxTemp turb timeBgn NA
## 29 data fluxTemp turb timeEnd NA
## 30 data fluxTemp turb flux W m-2
## 31 data foot stat timeBgn NA
## 32 data foot stat timeEnd NA
## 33 data foot stat angZaxsErth deg
## 34 data foot stat distReso m
## 35 data foot stat veloYaxsHorSd m s-1
## 36 data foot stat veloZaxsHorSd m s-1
## 37 data foot stat veloFric m s-1
## 38 data foot stat distZaxsMeasDisp m
## 39 data foot stat distZaxsRgh m
## 40 data foot stat distZaxsAbl m
## 41 data foot stat distXaxs90 m
## 42 data foot stat distXaxsMax m
## 43 data foot stat distYaxs90 m
## 44 qfqm fluxCo2 nsae timeBgn NA
## 45 qfqm fluxCo2 nsae timeEnd NA
## 46 qfqm fluxCo2 nsae qfFinl NA
## 47 qfqm fluxCo2 stor qfFinl NA
## 48 qfqm fluxCo2 stor timeBgn NA
## 49 qfqm fluxCo2 stor timeEnd NA
## 50 qfqm fluxCo2 turb timeBgn NA
## 51 qfqm fluxCo2 turb timeEnd NA
## 52 qfqm fluxCo2 turb qfFinl NA
## 53 qfqm fluxH2o nsae timeBgn NA
## 54 qfqm fluxH2o nsae timeEnd NA
## 55 qfqm fluxH2o nsae qfFinl NA
## 56 qfqm fluxH2o stor qfFinl NA
## 57 qfqm fluxH2o stor timeBgn NA
## 58 qfqm fluxH2o stor timeEnd NA
## 59 qfqm fluxH2o turb timeBgn NA
## 60 qfqm fluxH2o turb timeEnd NA
## 61 qfqm fluxH2o turb qfFinl NA
## 62 qfqm fluxMome turb timeBgn NA
## 63 qfqm fluxMome turb timeEnd NA
## 64 qfqm fluxMome turb qfFinl NA
## 65 qfqm fluxTemp nsae timeBgn NA
## 66 qfqm fluxTemp nsae timeEnd NA
## 67 qfqm fluxTemp nsae qfFinl NA
## 68 qfqm fluxTemp stor qfFinl NA
## 69 qfqm fluxTemp stor timeBgn NA
## 70 qfqm fluxTemp stor timeEnd NA
## 71 qfqm fluxTemp turb timeBgn NA
## 72 qfqm fluxTemp turb timeEnd NA
## 73 qfqm fluxTemp turb qfFinl NA
## 74 qfqm foot turb timeBgn NA
## 75 qfqm foot turb timeEnd NA
## 76 qfqm foot turb qfFinl NA
Let's plot some data! First, a brief aside about time stamps, since these are time series data.
Time stamps
NEON sensor data come with time stamps for both the start and end of the averaging period. Depending on the analysis you're doing, you may want to use one or the other; for general plotting, re-formatting, and transformations, I prefer to use the start time, because there are some small inconsistencies between data products in a few of the end time stamps.
Note that all NEON data use UTC time, aka Greenwich Mean Time.
This is true across NEON's instrumented, observational, and airborne
measurements. When working with NEON data, it's best to keep
everything in UTC as much as possible, otherwise it's very easy to
end up with data in mismatched times, which can cause insidious and
hard-to-detect problems. In the code below, time stamps and time
zones have been handled by stackEddy() and loadByProduct(), so we
don't need to do anything additional. But if you're writing your own
code and need to convert times, remember that if the time zone isn't
specified, R will default to the local time zone it detects on your
operating system.
plot(flux$NIWO$data.fluxCo2.nsae.flux~flux$NIWO$timeBgn,
pch=".", xlab="Date", ylab="CO2 flux")
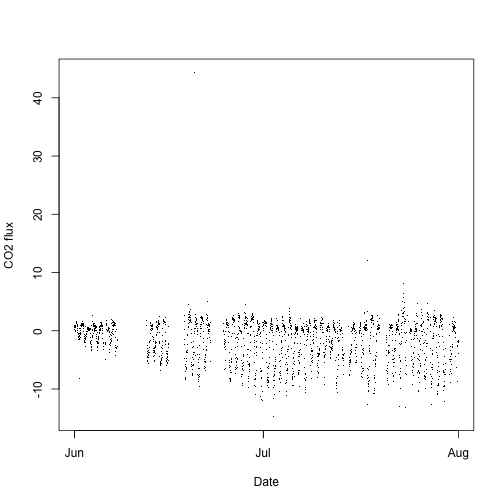
There is a clear diurnal pattern, and an increase in daily carbon uptake as the growing season progresses.
Let's trim down to just two days of data to see a few other details.
plot(flux$NIWO$data.fluxCo2.nsae.flux~flux$NIWO$timeBgn,
pch=20, xlab="Date", ylab="CO2 flux",
xlim=c(as.POSIXct("2018-07-07", tz="GMT"),
as.POSIXct("2018-07-09", tz="GMT")),
ylim=c(-20,20), xaxt="n")
axis.POSIXct(1, x=flux$NIWO$timeBgn,
format="%Y-%m-%d %H:%M:%S")
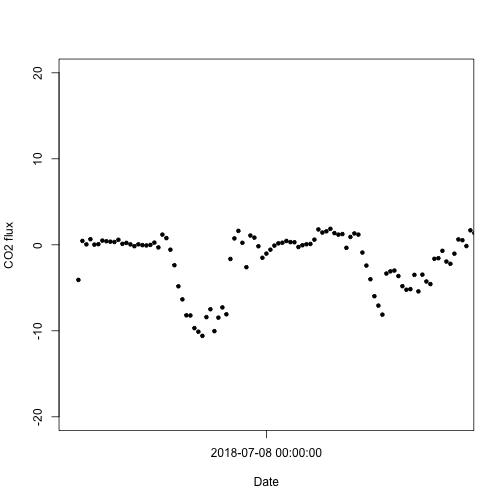
Note the timing of C uptake; the UTC time zone is clear here, where uptake occurs at times that appear to be during the night.
4. Merge flux data with other sensor data
Many of the data sets we would use to interpret and model flux data are measured as part of the NEON project, but are not present in the eddy flux data product bundle. In this section, we'll download PAR data and merge them with the flux data; the steps taken here can be applied to any of the NEON instrumented (IS) data products.
Download PAR data
To get NEON PAR data, use the loadByProduct() function from the
neonUtilities package. loadByProduct() takes the same inputs as
zipsByProduct(), but it loads the downloaded data directly into the
current R environment.
Let's download PAR data matching the Niwot Ridge flux data. The inputs needed are:
-
dpID: DP1.00024.001 -
site: NIWO -
startdate: 2018-06 -
enddate: 2018-07 -
package: basic -
timeIndex: 30
The new input here is timeIndex=30, which downloads only the 30-minute data.
Since the flux data are at a 30-minute resolution, we can save on
download time by disregarding the 1-minute data files (which are of course
30 times larger). The timeIndex input can be left off if you want to download
all available averaging intervals.
pr <- loadByProduct("DP1.00024.001", site="NIWO",
timeIndex=30, package="basic",
startdate="2018-06", enddate="2018-07",
check.size=F)
pr is another named list, and again, metadata and units can be found in
the variables table. The PARPAR_30min table contains a verticalPosition
field. This field indicates the position on the tower, with 10 being the
first tower level, and 20, 30, etc going up the tower.
Join PAR to flux data
We'll connect PAR data from the tower top to the flux data.
pr.top <- pr$PARPAR_30min[which(pr$PARPAR_30min$verticalPosition==
max(pr$PARPAR_30min$verticalPosition)),]
As noted above, loadByProduct() automatically converts time stamps
to a recognized date-time format when it reads the data. However, the
field names for the time stamps differ between the flux data and the
other meteorological data: the start of the averaging interval is
timeBgn in the flux data and startDateTime in the PAR data.
Let's create a new variable in the PAR data:
pr.top$timeBgn <- pr.top$startDateTime
And now use the matching time stamp fields to merge the flux and PAR data.
fx.pr <- merge(pr.top, flux$NIWO, by="timeBgn")
And now we can plot net carbon exchange as a function of light availability:
plot(fx.pr$data.fluxCo2.nsae.flux~fx.pr$PARMean,
pch=".", ylim=c(-20,20),
xlab="PAR", ylab="CO2 flux")
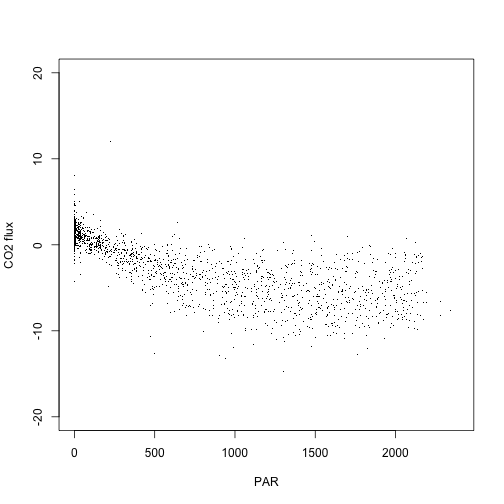
If you're interested in data in the eddy covariance bundle besides the net flux data, the rest of this tutorial will guide you through how to get those data out of the bundle.
5. Vertical profile data (Level 3)
The Level 3 (dp03) data are the spatially interpolated profiles of
the rates of change of CO2, H2O, and temperature.
Extract the Level 3 data from the HDF5 file using stackEddy() with
the same syntax as for the Level 4 data.
prof <- stackEddy(filepath="~/Downloads/filesToStack00200/",
level="dp03")
As with the Level 4 data, the result is a named list with data tables for each site.
head(prof$NIWO)
## timeBgn timeEnd data.co2Stor.rateRtioMoleDryCo2.X0.1.m data.co2Stor.rateRtioMoleDryCo2.X0.2.m
## 1 2018-06-01 2018-06-01 00:29:59 -0.0002681938 -0.0002681938
## data.co2Stor.rateRtioMoleDryCo2.X0.3.m data.co2Stor.rateRtioMoleDryCo2.X0.4.m
## 1 -0.0002681938 -0.0002681938
## data.co2Stor.rateRtioMoleDryCo2.X0.5.m data.co2Stor.rateRtioMoleDryCo2.X0.6.m
## 1 -0.0002681938 -0.0002681938
## data.co2Stor.rateRtioMoleDryCo2.X0.7.m data.co2Stor.rateRtioMoleDryCo2.X0.8.m
## 1 -0.0002681938 -0.0002681938
## data.co2Stor.rateRtioMoleDryCo2.X0.9.m data.co2Stor.rateRtioMoleDryCo2.X1.m
## 1 -0.0002681938 -0.0002681938
## data.co2Stor.rateRtioMoleDryCo2.X1.1.m data.co2Stor.rateRtioMoleDryCo2.X1.2.m
## 1 -0.0002681938 -0.0002681938
## data.co2Stor.rateRtioMoleDryCo2.X1.3.m data.co2Stor.rateRtioMoleDryCo2.X1.4.m
## 1 -0.0002681938 -0.0002681938
## data.co2Stor.rateRtioMoleDryCo2.X1.5.m data.co2Stor.rateRtioMoleDryCo2.X1.6.m
## 1 -0.0002681938 -0.0002681938
## data.co2Stor.rateRtioMoleDryCo2.X1.7.m data.co2Stor.rateRtioMoleDryCo2.X1.8.m
## 1 -0.0002681938 -0.0002681938
## data.co2Stor.rateRtioMoleDryCo2.X1.9.m data.co2Stor.rateRtioMoleDryCo2.X2.m
## 1 -0.0002681938 -0.0002681938
## data.co2Stor.rateRtioMoleDryCo2.X2.1.m data.co2Stor.rateRtioMoleDryCo2.X2.2.m
## 1 -0.0002681938 -0.0002681938
## data.co2Stor.rateRtioMoleDryCo2.X2.3.m data.co2Stor.rateRtioMoleDryCo2.X2.4.m
## 1 -0.0002681938 -0.0002681938
## data.co2Stor.rateRtioMoleDryCo2.X2.5.m data.co2Stor.rateRtioMoleDryCo2.X2.6.m
## 1 -0.0002681938 -0.0002681938
## data.co2Stor.rateRtioMoleDryCo2.X2.7.m data.co2Stor.rateRtioMoleDryCo2.X2.8.m
## 1 -0.0002681938 -0.0002681938
## data.co2Stor.rateRtioMoleDryCo2.X2.9.m data.co2Stor.rateRtioMoleDryCo2.X3.m
## 1 -0.0002681938 -0.0002681938
## data.co2Stor.rateRtioMoleDryCo2.X3.1.m data.co2Stor.rateRtioMoleDryCo2.X3.2.m
## 1 -0.0002681938 -0.0002681938
## data.co2Stor.rateRtioMoleDryCo2.X3.3.m data.co2Stor.rateRtioMoleDryCo2.X3.4.m
## 1 -0.0002681938 -0.0002681938
## data.co2Stor.rateRtioMoleDryCo2.X3.5.m data.co2Stor.rateRtioMoleDryCo2.X3.6.m
## 1 -0.0002681938 -0.0002681938
## data.co2Stor.rateRtioMoleDryCo2.X3.7.m data.co2Stor.rateRtioMoleDryCo2.X3.8.m
## 1 -0.0002681938 -0.0002681938
## data.co2Stor.rateRtioMoleDryCo2.X3.9.m data.co2Stor.rateRtioMoleDryCo2.X4.m
## 1 -0.0002681938 -0.0002681938
## data.co2Stor.rateRtioMoleDryCo2.X4.1.m data.co2Stor.rateRtioMoleDryCo2.X4.2.m
## 1 -0.0002681938 -0.0002681938
## data.co2Stor.rateRtioMoleDryCo2.X4.3.m data.co2Stor.rateRtioMoleDryCo2.X4.4.m
## 1 -0.0002681938 -0.0002681938
## data.co2Stor.rateRtioMoleDryCo2.X4.5.m data.co2Stor.rateRtioMoleDryCo2.X4.6.m
## 1 -0.0002681938 -0.0002681938
## data.co2Stor.rateRtioMoleDryCo2.X4.7.m data.co2Stor.rateRtioMoleDryCo2.X4.8.m
## 1 -0.0002681938 -0.0002681938
## data.co2Stor.rateRtioMoleDryCo2.X4.9.m data.co2Stor.rateRtioMoleDryCo2.X5.m
## 1 -0.0002681938 -0.0002681938
## data.co2Stor.rateRtioMoleDryCo2.X5.1.m data.co2Stor.rateRtioMoleDryCo2.X5.2.m
## 1 -0.0002681938 -0.0002681938
## data.co2Stor.rateRtioMoleDryCo2.X5.3.m data.co2Stor.rateRtioMoleDryCo2.X5.4.m
## 1 -0.0002681938 -0.0002681938
## data.co2Stor.rateRtioMoleDryCo2.X5.5.m data.co2Stor.rateRtioMoleDryCo2.X5.6.m
## 1 -0.0002681938 -0.0002681938
## data.co2Stor.rateRtioMoleDryCo2.X5.7.m data.co2Stor.rateRtioMoleDryCo2.X5.8.m
## 1 -0.0002681938 -0.0002681938
## data.co2Stor.rateRtioMoleDryCo2.X5.9.m data.co2Stor.rateRtioMoleDryCo2.X6.m
## 1 -0.0002681938 -0.0002681938
## data.co2Stor.rateRtioMoleDryCo2.X6.1.m data.co2Stor.rateRtioMoleDryCo2.X6.2.m
## 1 -0.0002681938 -0.0002681938
## data.co2Stor.rateRtioMoleDryCo2.X6.3.m data.co2Stor.rateRtioMoleDryCo2.X6.4.m
## 1 -0.0002681938 -0.0002681938
## data.co2Stor.rateRtioMoleDryCo2.X6.5.m data.co2Stor.rateRtioMoleDryCo2.X6.6.m
## 1 -0.0002681938 -0.0002681938
## data.co2Stor.rateRtioMoleDryCo2.X6.7.m data.co2Stor.rateRtioMoleDryCo2.X6.8.m
## 1 -0.0002681938 -0.0002681938
## data.co2Stor.rateRtioMoleDryCo2.X6.9.m data.co2Stor.rateRtioMoleDryCo2.X7.m
## 1 -0.0002681938 -0.0002681938
## data.co2Stor.rateRtioMoleDryCo2.X7.1.m data.co2Stor.rateRtioMoleDryCo2.X7.2.m
## 1 -0.0002681938 -0.0002681938
## data.co2Stor.rateRtioMoleDryCo2.X7.3.m data.co2Stor.rateRtioMoleDryCo2.X7.4.m
## 1 -0.0002681938 -0.0002681938
## data.co2Stor.rateRtioMoleDryCo2.X7.5.m data.co2Stor.rateRtioMoleDryCo2.X7.6.m
## 1 -0.0002681938 -0.0002681938
## data.co2Stor.rateRtioMoleDryCo2.X7.7.m data.co2Stor.rateRtioMoleDryCo2.X7.8.m
## 1 -0.0002681938 -0.0002681938
## data.co2Stor.rateRtioMoleDryCo2.X7.9.m data.co2Stor.rateRtioMoleDryCo2.X8.m
## 1 -0.0002681938 -0.0002681938
## data.co2Stor.rateRtioMoleDryCo2.X8.1.m data.co2Stor.rateRtioMoleDryCo2.X8.2.m
## 1 -0.0002681938 -0.0002681938
## data.co2Stor.rateRtioMoleDryCo2.X8.3.m data.co2Stor.rateRtioMoleDryCo2.X8.4.m
## 1 -0.0002681938 -0.0002681938
## data.h2oStor.rateRtioMoleDryH2o.X0.1.m data.h2oStor.rateRtioMoleDryH2o.X0.2.m
## 1 0.000315911 0.000315911
## data.h2oStor.rateRtioMoleDryH2o.X0.3.m data.h2oStor.rateRtioMoleDryH2o.X0.4.m
## 1 0.000315911 0.000315911
## data.h2oStor.rateRtioMoleDryH2o.X0.5.m data.h2oStor.rateRtioMoleDryH2o.X0.6.m
## 1 0.000315911 0.000315911
## data.h2oStor.rateRtioMoleDryH2o.X0.7.m data.h2oStor.rateRtioMoleDryH2o.X0.8.m
## 1 0.000315911 0.000315911
## data.h2oStor.rateRtioMoleDryH2o.X0.9.m data.h2oStor.rateRtioMoleDryH2o.X1.m
## 1 0.000315911 0.000315911
## data.h2oStor.rateRtioMoleDryH2o.X1.1.m data.h2oStor.rateRtioMoleDryH2o.X1.2.m
## 1 0.000315911 0.000315911
## data.h2oStor.rateRtioMoleDryH2o.X1.3.m data.h2oStor.rateRtioMoleDryH2o.X1.4.m
## 1 0.000315911 0.000315911
## data.h2oStor.rateRtioMoleDryH2o.X1.5.m data.h2oStor.rateRtioMoleDryH2o.X1.6.m
## 1 0.000315911 0.000315911
## data.h2oStor.rateRtioMoleDryH2o.X1.7.m data.h2oStor.rateRtioMoleDryH2o.X1.8.m
## 1 0.000315911 0.000315911
## data.h2oStor.rateRtioMoleDryH2o.X1.9.m data.h2oStor.rateRtioMoleDryH2o.X2.m
## 1 0.000315911 0.000315911
## data.h2oStor.rateRtioMoleDryH2o.X2.1.m data.h2oStor.rateRtioMoleDryH2o.X2.2.m
## 1 0.000315911 0.000315911
## data.h2oStor.rateRtioMoleDryH2o.X2.3.m data.h2oStor.rateRtioMoleDryH2o.X2.4.m
## 1 0.000315911 0.000315911
## data.h2oStor.rateRtioMoleDryH2o.X2.5.m data.h2oStor.rateRtioMoleDryH2o.X2.6.m
## 1 0.000315911 0.000315911
## data.h2oStor.rateRtioMoleDryH2o.X2.7.m data.h2oStor.rateRtioMoleDryH2o.X2.8.m
## 1 0.000315911 0.000315911
## data.h2oStor.rateRtioMoleDryH2o.X2.9.m data.h2oStor.rateRtioMoleDryH2o.X3.m
## 1 0.000315911 0.000315911
## data.h2oStor.rateRtioMoleDryH2o.X3.1.m data.h2oStor.rateRtioMoleDryH2o.X3.2.m
## 1 0.000315911 0.000315911
## data.h2oStor.rateRtioMoleDryH2o.X3.3.m data.h2oStor.rateRtioMoleDryH2o.X3.4.m
## 1 0.000315911 0.000315911
## data.h2oStor.rateRtioMoleDryH2o.X3.5.m data.h2oStor.rateRtioMoleDryH2o.X3.6.m
## 1 0.000315911 0.000315911
## data.h2oStor.rateRtioMoleDryH2o.X3.7.m data.h2oStor.rateRtioMoleDryH2o.X3.8.m
## 1 0.000315911 0.000315911
## data.h2oStor.rateRtioMoleDryH2o.X3.9.m data.h2oStor.rateRtioMoleDryH2o.X4.m
## 1 0.000315911 0.000315911
## data.h2oStor.rateRtioMoleDryH2o.X4.1.m data.h2oStor.rateRtioMoleDryH2o.X4.2.m
## 1 0.000315911 0.000315911
## data.h2oStor.rateRtioMoleDryH2o.X4.3.m data.h2oStor.rateRtioMoleDryH2o.X4.4.m
## 1 0.000315911 0.000315911
## data.h2oStor.rateRtioMoleDryH2o.X4.5.m data.h2oStor.rateRtioMoleDryH2o.X4.6.m
## 1 0.000315911 0.000315911
## data.h2oStor.rateRtioMoleDryH2o.X4.7.m data.h2oStor.rateRtioMoleDryH2o.X4.8.m
## 1 0.000315911 0.000315911
## data.h2oStor.rateRtioMoleDryH2o.X4.9.m data.h2oStor.rateRtioMoleDryH2o.X5.m
## 1 0.000315911 0.000315911
## data.h2oStor.rateRtioMoleDryH2o.X5.1.m data.h2oStor.rateRtioMoleDryH2o.X5.2.m
## 1 0.000315911 0.000315911
## data.h2oStor.rateRtioMoleDryH2o.X5.3.m data.h2oStor.rateRtioMoleDryH2o.X5.4.m
## 1 0.000315911 0.000315911
## data.h2oStor.rateRtioMoleDryH2o.X5.5.m data.h2oStor.rateRtioMoleDryH2o.X5.6.m
## 1 0.000315911 0.000315911
## data.h2oStor.rateRtioMoleDryH2o.X5.7.m data.h2oStor.rateRtioMoleDryH2o.X5.8.m
## 1 0.000315911 0.000315911
## data.h2oStor.rateRtioMoleDryH2o.X5.9.m data.h2oStor.rateRtioMoleDryH2o.X6.m
## 1 0.000315911 0.000315911
## data.h2oStor.rateRtioMoleDryH2o.X6.1.m data.h2oStor.rateRtioMoleDryH2o.X6.2.m
## 1 0.000315911 0.000315911
## data.h2oStor.rateRtioMoleDryH2o.X6.3.m data.h2oStor.rateRtioMoleDryH2o.X6.4.m
## 1 0.000315911 0.000315911
## data.h2oStor.rateRtioMoleDryH2o.X6.5.m data.h2oStor.rateRtioMoleDryH2o.X6.6.m
## 1 0.000315911 0.000315911
## data.h2oStor.rateRtioMoleDryH2o.X6.7.m data.h2oStor.rateRtioMoleDryH2o.X6.8.m
## 1 0.000315911 0.000315911
## data.h2oStor.rateRtioMoleDryH2o.X6.9.m data.h2oStor.rateRtioMoleDryH2o.X7.m
## 1 0.000315911 0.000315911
## data.h2oStor.rateRtioMoleDryH2o.X7.1.m data.h2oStor.rateRtioMoleDryH2o.X7.2.m
## 1 0.000315911 0.000315911
## data.h2oStor.rateRtioMoleDryH2o.X7.3.m data.h2oStor.rateRtioMoleDryH2o.X7.4.m
## 1 0.000315911 0.000315911
## data.h2oStor.rateRtioMoleDryH2o.X7.5.m data.h2oStor.rateRtioMoleDryH2o.X7.6.m
## 1 0.000315911 0.000315911
## data.h2oStor.rateRtioMoleDryH2o.X7.7.m data.h2oStor.rateRtioMoleDryH2o.X7.8.m
## 1 0.000315911 0.000315911
## data.h2oStor.rateRtioMoleDryH2o.X7.9.m data.h2oStor.rateRtioMoleDryH2o.X8.m
## 1 0.000315911 0.000315911
## data.h2oStor.rateRtioMoleDryH2o.X8.1.m data.h2oStor.rateRtioMoleDryH2o.X8.2.m
## 1 0.000315911 0.000315911
## data.h2oStor.rateRtioMoleDryH2o.X8.3.m data.h2oStor.rateRtioMoleDryH2o.X8.4.m data.tempStor.rateTemp.X0.1.m
## 1 0.000315911 0.000315911 -0.0001014444
## data.tempStor.rateTemp.X0.2.m data.tempStor.rateTemp.X0.3.m data.tempStor.rateTemp.X0.4.m
## 1 -0.0001014444 -0.0001014444 -0.0001014444
## data.tempStor.rateTemp.X0.5.m data.tempStor.rateTemp.X0.6.m data.tempStor.rateTemp.X0.7.m
## 1 -0.0001014444 -0.0001050874 -0.000111159
## data.tempStor.rateTemp.X0.8.m data.tempStor.rateTemp.X0.9.m data.tempStor.rateTemp.X1.m
## 1 -0.0001172305 -0.0001233021 -0.0001293737
## data.tempStor.rateTemp.X1.1.m data.tempStor.rateTemp.X1.2.m data.tempStor.rateTemp.X1.3.m
## 1 -0.0001354453 -0.0001415168 -0.0001475884
## data.tempStor.rateTemp.X1.4.m data.tempStor.rateTemp.X1.5.m data.tempStor.rateTemp.X1.6.m
## 1 -0.00015366 -0.0001597315 -0.0001658031
## data.tempStor.rateTemp.X1.7.m data.tempStor.rateTemp.X1.8.m data.tempStor.rateTemp.X1.9.m
## 1 -0.0001718747 -0.0001779463 -0.0001840178
## data.tempStor.rateTemp.X2.m data.tempStor.rateTemp.X2.1.m data.tempStor.rateTemp.X2.2.m
## 1 -0.000185739 -0.0001869767 -0.0001882144
## data.tempStor.rateTemp.X2.3.m data.tempStor.rateTemp.X2.4.m data.tempStor.rateTemp.X2.5.m
## 1 -0.0001894521 -0.0001906899 -0.0001919276
## data.tempStor.rateTemp.X2.6.m data.tempStor.rateTemp.X2.7.m data.tempStor.rateTemp.X2.8.m
## 1 -0.0001931653 -0.0001944031 -0.0001956408
## data.tempStor.rateTemp.X2.9.m data.tempStor.rateTemp.X3.m data.tempStor.rateTemp.X3.1.m
## 1 -0.0001968785 -0.0001981162 -0.000199354
## data.tempStor.rateTemp.X3.2.m data.tempStor.rateTemp.X3.3.m data.tempStor.rateTemp.X3.4.m
## 1 -0.0002005917 -0.0002018294 -0.0002030672
## data.tempStor.rateTemp.X3.5.m data.tempStor.rateTemp.X3.6.m data.tempStor.rateTemp.X3.7.m
## 1 -0.0002043049 -0.0002055426 -0.0002067803
## data.tempStor.rateTemp.X3.8.m data.tempStor.rateTemp.X3.9.m data.tempStor.rateTemp.X4.m
## 1 -0.0002080181 -0.0002092558 -0.0002104935
## data.tempStor.rateTemp.X4.1.m data.tempStor.rateTemp.X4.2.m data.tempStor.rateTemp.X4.3.m
## 1 -0.0002117313 -0.000212969 -0.0002142067
## data.tempStor.rateTemp.X4.4.m data.tempStor.rateTemp.X4.5.m data.tempStor.rateTemp.X4.6.m
## 1 -0.0002154444 -0.0002172161 -0.0002189878
## data.tempStor.rateTemp.X4.7.m data.tempStor.rateTemp.X4.8.m data.tempStor.rateTemp.X4.9.m
## 1 -0.0002207595 -0.0002225312 -0.0002243029
## data.tempStor.rateTemp.X5.m data.tempStor.rateTemp.X5.1.m data.tempStor.rateTemp.X5.2.m
## 1 -0.0002260746 -0.0002278463 -0.000229618
## data.tempStor.rateTemp.X5.3.m data.tempStor.rateTemp.X5.4.m data.tempStor.rateTemp.X5.5.m
## 1 -0.0002313896 -0.0002331613 -0.000234933
## data.tempStor.rateTemp.X5.6.m data.tempStor.rateTemp.X5.7.m data.tempStor.rateTemp.X5.8.m
## 1 -0.0002367047 -0.0002384764 -0.0002402481
## data.tempStor.rateTemp.X5.9.m data.tempStor.rateTemp.X6.m data.tempStor.rateTemp.X6.1.m
## 1 -0.0002420198 -0.0002437915 -0.0002455631
## data.tempStor.rateTemp.X6.2.m data.tempStor.rateTemp.X6.3.m data.tempStor.rateTemp.X6.4.m
## 1 -0.0002473348 -0.0002491065 -0.0002508782
## data.tempStor.rateTemp.X6.5.m data.tempStor.rateTemp.X6.6.m data.tempStor.rateTemp.X6.7.m
## 1 -0.0002526499 -0.0002544216 -0.0002561933
## data.tempStor.rateTemp.X6.8.m data.tempStor.rateTemp.X6.9.m data.tempStor.rateTemp.X7.m
## 1 -0.000257965 -0.0002597367 -0.0002615083
## data.tempStor.rateTemp.X7.1.m data.tempStor.rateTemp.X7.2.m data.tempStor.rateTemp.X7.3.m
## 1 -0.00026328 -0.0002650517 -0.0002668234
## data.tempStor.rateTemp.X7.4.m data.tempStor.rateTemp.X7.5.m data.tempStor.rateTemp.X7.6.m
## 1 -0.0002685951 -0.0002703668 -0.0002721385
## data.tempStor.rateTemp.X7.7.m data.tempStor.rateTemp.X7.8.m data.tempStor.rateTemp.X7.9.m
## 1 -0.0002739102 -0.0002756819 -0.0002774535
## data.tempStor.rateTemp.X8.m data.tempStor.rateTemp.X8.1.m data.tempStor.rateTemp.X8.2.m
## 1 -0.0002792252 -0.0002809969 -0.0002827686
## data.tempStor.rateTemp.X8.3.m data.tempStor.rateTemp.X8.4.m qfqm.co2Stor.rateRtioMoleDryCo2.X0.1.m
## 1 -0.0002845403 -0.000286312 1
## qfqm.co2Stor.rateRtioMoleDryCo2.X0.2.m qfqm.co2Stor.rateRtioMoleDryCo2.X0.3.m
## 1 1 1
## qfqm.co2Stor.rateRtioMoleDryCo2.X0.4.m qfqm.co2Stor.rateRtioMoleDryCo2.X0.5.m
## 1 1 1
## qfqm.co2Stor.rateRtioMoleDryCo2.X0.6.m qfqm.co2Stor.rateRtioMoleDryCo2.X0.7.m
## 1 1 1
## qfqm.co2Stor.rateRtioMoleDryCo2.X0.8.m qfqm.co2Stor.rateRtioMoleDryCo2.X0.9.m
## 1 1 1
## qfqm.co2Stor.rateRtioMoleDryCo2.X1.m qfqm.co2Stor.rateRtioMoleDryCo2.X1.1.m
## 1 1 1
## qfqm.co2Stor.rateRtioMoleDryCo2.X1.2.m qfqm.co2Stor.rateRtioMoleDryCo2.X1.3.m
## 1 1 1
## qfqm.co2Stor.rateRtioMoleDryCo2.X1.4.m qfqm.co2Stor.rateRtioMoleDryCo2.X1.5.m
## 1 1 1
## qfqm.co2Stor.rateRtioMoleDryCo2.X1.6.m qfqm.co2Stor.rateRtioMoleDryCo2.X1.7.m
## 1 1 1
## qfqm.co2Stor.rateRtioMoleDryCo2.X1.8.m qfqm.co2Stor.rateRtioMoleDryCo2.X1.9.m
## 1 1 1
## qfqm.co2Stor.rateRtioMoleDryCo2.X2.m qfqm.co2Stor.rateRtioMoleDryCo2.X2.1.m
## 1 1 1
## qfqm.co2Stor.rateRtioMoleDryCo2.X2.2.m qfqm.co2Stor.rateRtioMoleDryCo2.X2.3.m
## 1 1 1
## qfqm.co2Stor.rateRtioMoleDryCo2.X2.4.m qfqm.co2Stor.rateRtioMoleDryCo2.X2.5.m
## 1 1 1
## qfqm.co2Stor.rateRtioMoleDryCo2.X2.6.m qfqm.co2Stor.rateRtioMoleDryCo2.X2.7.m
## 1 1 1
## qfqm.co2Stor.rateRtioMoleDryCo2.X2.8.m qfqm.co2Stor.rateRtioMoleDryCo2.X2.9.m
## 1 1 1
## qfqm.co2Stor.rateRtioMoleDryCo2.X3.m qfqm.co2Stor.rateRtioMoleDryCo2.X3.1.m
## 1 1 1
## qfqm.co2Stor.rateRtioMoleDryCo2.X3.2.m qfqm.co2Stor.rateRtioMoleDryCo2.X3.3.m
## 1 1 1
## qfqm.co2Stor.rateRtioMoleDryCo2.X3.4.m qfqm.co2Stor.rateRtioMoleDryCo2.X3.5.m
## 1 1 1
## qfqm.co2Stor.rateRtioMoleDryCo2.X3.6.m qfqm.co2Stor.rateRtioMoleDryCo2.X3.7.m
## 1 1 1
## qfqm.co2Stor.rateRtioMoleDryCo2.X3.8.m qfqm.co2Stor.rateRtioMoleDryCo2.X3.9.m
## 1 1 1
## qfqm.co2Stor.rateRtioMoleDryCo2.X4.m qfqm.co2Stor.rateRtioMoleDryCo2.X4.1.m
## 1 1 1
## qfqm.co2Stor.rateRtioMoleDryCo2.X4.2.m qfqm.co2Stor.rateRtioMoleDryCo2.X4.3.m
## 1 1 1
## qfqm.co2Stor.rateRtioMoleDryCo2.X4.4.m qfqm.co2Stor.rateRtioMoleDryCo2.X4.5.m
## 1 1 1
## qfqm.co2Stor.rateRtioMoleDryCo2.X4.6.m qfqm.co2Stor.rateRtioMoleDryCo2.X4.7.m
## 1 1 1
## qfqm.co2Stor.rateRtioMoleDryCo2.X4.8.m qfqm.co2Stor.rateRtioMoleDryCo2.X4.9.m
## 1 1 1
## qfqm.co2Stor.rateRtioMoleDryCo2.X5.m qfqm.co2Stor.rateRtioMoleDryCo2.X5.1.m
## 1 1 1
## qfqm.co2Stor.rateRtioMoleDryCo2.X5.2.m qfqm.co2Stor.rateRtioMoleDryCo2.X5.3.m
## 1 1 1
## qfqm.co2Stor.rateRtioMoleDryCo2.X5.4.m qfqm.co2Stor.rateRtioMoleDryCo2.X5.5.m
## 1 1 1
## qfqm.co2Stor.rateRtioMoleDryCo2.X5.6.m qfqm.co2Stor.rateRtioMoleDryCo2.X5.7.m
## 1 1 1
## qfqm.co2Stor.rateRtioMoleDryCo2.X5.8.m qfqm.co2Stor.rateRtioMoleDryCo2.X5.9.m
## 1 1 1
## qfqm.co2Stor.rateRtioMoleDryCo2.X6.m qfqm.co2Stor.rateRtioMoleDryCo2.X6.1.m
## 1 1 1
## qfqm.co2Stor.rateRtioMoleDryCo2.X6.2.m qfqm.co2Stor.rateRtioMoleDryCo2.X6.3.m
## 1 1 1
## qfqm.co2Stor.rateRtioMoleDryCo2.X6.4.m qfqm.co2Stor.rateRtioMoleDryCo2.X6.5.m
## 1 1 1
## qfqm.co2Stor.rateRtioMoleDryCo2.X6.6.m qfqm.co2Stor.rateRtioMoleDryCo2.X6.7.m
## 1 1 1
## qfqm.co2Stor.rateRtioMoleDryCo2.X6.8.m qfqm.co2Stor.rateRtioMoleDryCo2.X6.9.m
## 1 1 1
## qfqm.co2Stor.rateRtioMoleDryCo2.X7.m qfqm.co2Stor.rateRtioMoleDryCo2.X7.1.m
## 1 1 1
## qfqm.co2Stor.rateRtioMoleDryCo2.X7.2.m qfqm.co2Stor.rateRtioMoleDryCo2.X7.3.m
## 1 1 1
## qfqm.co2Stor.rateRtioMoleDryCo2.X7.4.m qfqm.co2Stor.rateRtioMoleDryCo2.X7.5.m
## 1 1 1
## qfqm.co2Stor.rateRtioMoleDryCo2.X7.6.m qfqm.co2Stor.rateRtioMoleDryCo2.X7.7.m
## 1 1 1
## qfqm.co2Stor.rateRtioMoleDryCo2.X7.8.m qfqm.co2Stor.rateRtioMoleDryCo2.X7.9.m
## 1 1 1
## qfqm.co2Stor.rateRtioMoleDryCo2.X8.m qfqm.co2Stor.rateRtioMoleDryCo2.X8.1.m
## 1 1 1
## qfqm.co2Stor.rateRtioMoleDryCo2.X8.2.m qfqm.co2Stor.rateRtioMoleDryCo2.X8.3.m
## 1 1 1
## qfqm.co2Stor.rateRtioMoleDryCo2.X8.4.m qfqm.h2oStor.rateRtioMoleDryH2o.X0.1.m
## 1 1 1
## qfqm.h2oStor.rateRtioMoleDryH2o.X0.2.m qfqm.h2oStor.rateRtioMoleDryH2o.X0.3.m
## 1 1 1
## qfqm.h2oStor.rateRtioMoleDryH2o.X0.4.m qfqm.h2oStor.rateRtioMoleDryH2o.X0.5.m
## 1 1 1
## qfqm.h2oStor.rateRtioMoleDryH2o.X0.6.m qfqm.h2oStor.rateRtioMoleDryH2o.X0.7.m
## 1 1 1
## qfqm.h2oStor.rateRtioMoleDryH2o.X0.8.m qfqm.h2oStor.rateRtioMoleDryH2o.X0.9.m
## 1 1 1
## qfqm.h2oStor.rateRtioMoleDryH2o.X1.m qfqm.h2oStor.rateRtioMoleDryH2o.X1.1.m
## 1 1 1
## qfqm.h2oStor.rateRtioMoleDryH2o.X1.2.m qfqm.h2oStor.rateRtioMoleDryH2o.X1.3.m
## 1 1 1
## qfqm.h2oStor.rateRtioMoleDryH2o.X1.4.m qfqm.h2oStor.rateRtioMoleDryH2o.X1.5.m
## 1 1 1
## qfqm.h2oStor.rateRtioMoleDryH2o.X1.6.m qfqm.h2oStor.rateRtioMoleDryH2o.X1.7.m
## 1 1 1
## qfqm.h2oStor.rateRtioMoleDryH2o.X1.8.m qfqm.h2oStor.rateRtioMoleDryH2o.X1.9.m
## 1 1 1
## qfqm.h2oStor.rateRtioMoleDryH2o.X2.m qfqm.h2oStor.rateRtioMoleDryH2o.X2.1.m
## 1 1 1
## qfqm.h2oStor.rateRtioMoleDryH2o.X2.2.m qfqm.h2oStor.rateRtioMoleDryH2o.X2.3.m
## 1 1 1
## qfqm.h2oStor.rateRtioMoleDryH2o.X2.4.m qfqm.h2oStor.rateRtioMoleDryH2o.X2.5.m
## 1 1 1
## qfqm.h2oStor.rateRtioMoleDryH2o.X2.6.m qfqm.h2oStor.rateRtioMoleDryH2o.X2.7.m
## 1 1 1
## qfqm.h2oStor.rateRtioMoleDryH2o.X2.8.m qfqm.h2oStor.rateRtioMoleDryH2o.X2.9.m
## 1 1 1
## qfqm.h2oStor.rateRtioMoleDryH2o.X3.m qfqm.h2oStor.rateRtioMoleDryH2o.X3.1.m
## 1 1 1
## qfqm.h2oStor.rateRtioMoleDryH2o.X3.2.m qfqm.h2oStor.rateRtioMoleDryH2o.X3.3.m
## 1 1 1
## qfqm.h2oStor.rateRtioMoleDryH2o.X3.4.m qfqm.h2oStor.rateRtioMoleDryH2o.X3.5.m
## 1 1 1
## qfqm.h2oStor.rateRtioMoleDryH2o.X3.6.m qfqm.h2oStor.rateRtioMoleDryH2o.X3.7.m
## 1 1 1
## qfqm.h2oStor.rateRtioMoleDryH2o.X3.8.m qfqm.h2oStor.rateRtioMoleDryH2o.X3.9.m
## 1 1 1
## qfqm.h2oStor.rateRtioMoleDryH2o.X4.m qfqm.h2oStor.rateRtioMoleDryH2o.X4.1.m
## 1 1 1
## qfqm.h2oStor.rateRtioMoleDryH2o.X4.2.m qfqm.h2oStor.rateRtioMoleDryH2o.X4.3.m
## 1 1 1
## qfqm.h2oStor.rateRtioMoleDryH2o.X4.4.m qfqm.h2oStor.rateRtioMoleDryH2o.X4.5.m
## 1 1 1
## qfqm.h2oStor.rateRtioMoleDryH2o.X4.6.m qfqm.h2oStor.rateRtioMoleDryH2o.X4.7.m
## 1 1 1
## qfqm.h2oStor.rateRtioMoleDryH2o.X4.8.m qfqm.h2oStor.rateRtioMoleDryH2o.X4.9.m
## 1 1 1
## qfqm.h2oStor.rateRtioMoleDryH2o.X5.m qfqm.h2oStor.rateRtioMoleDryH2o.X5.1.m
## 1 1 1
## qfqm.h2oStor.rateRtioMoleDryH2o.X5.2.m qfqm.h2oStor.rateRtioMoleDryH2o.X5.3.m
## 1 1 1
## qfqm.h2oStor.rateRtioMoleDryH2o.X5.4.m qfqm.h2oStor.rateRtioMoleDryH2o.X5.5.m
## 1 1 1
## qfqm.h2oStor.rateRtioMoleDryH2o.X5.6.m qfqm.h2oStor.rateRtioMoleDryH2o.X5.7.m
## 1 1 1
## qfqm.h2oStor.rateRtioMoleDryH2o.X5.8.m qfqm.h2oStor.rateRtioMoleDryH2o.X5.9.m
## 1 1 1
## qfqm.h2oStor.rateRtioMoleDryH2o.X6.m qfqm.h2oStor.rateRtioMoleDryH2o.X6.1.m
## 1 1 1
## qfqm.h2oStor.rateRtioMoleDryH2o.X6.2.m qfqm.h2oStor.rateRtioMoleDryH2o.X6.3.m
## 1 1 1
## qfqm.h2oStor.rateRtioMoleDryH2o.X6.4.m qfqm.h2oStor.rateRtioMoleDryH2o.X6.5.m
## 1 1 1
## qfqm.h2oStor.rateRtioMoleDryH2o.X6.6.m qfqm.h2oStor.rateRtioMoleDryH2o.X6.7.m
## 1 1 1
## qfqm.h2oStor.rateRtioMoleDryH2o.X6.8.m qfqm.h2oStor.rateRtioMoleDryH2o.X6.9.m
## 1 1 1
## qfqm.h2oStor.rateRtioMoleDryH2o.X7.m qfqm.h2oStor.rateRtioMoleDryH2o.X7.1.m
## 1 1 1
## qfqm.h2oStor.rateRtioMoleDryH2o.X7.2.m qfqm.h2oStor.rateRtioMoleDryH2o.X7.3.m
## 1 1 1
## qfqm.h2oStor.rateRtioMoleDryH2o.X7.4.m qfqm.h2oStor.rateRtioMoleDryH2o.X7.5.m
## 1 1 1
## qfqm.h2oStor.rateRtioMoleDryH2o.X7.6.m qfqm.h2oStor.rateRtioMoleDryH2o.X7.7.m
## 1 1 1
## qfqm.h2oStor.rateRtioMoleDryH2o.X7.8.m qfqm.h2oStor.rateRtioMoleDryH2o.X7.9.m
## 1 1 1
## qfqm.h2oStor.rateRtioMoleDryH2o.X8.m qfqm.h2oStor.rateRtioMoleDryH2o.X8.1.m
## 1 1 1
## qfqm.h2oStor.rateRtioMoleDryH2o.X8.2.m qfqm.h2oStor.rateRtioMoleDryH2o.X8.3.m
## 1 1 1
## qfqm.h2oStor.rateRtioMoleDryH2o.X8.4.m qfqm.tempStor.rateTemp.X0.1.m qfqm.tempStor.rateTemp.X0.2.m
## 1 1 0 0
## qfqm.tempStor.rateTemp.X0.3.m qfqm.tempStor.rateTemp.X0.4.m qfqm.tempStor.rateTemp.X0.5.m
## 1 0 0 0
## qfqm.tempStor.rateTemp.X0.6.m qfqm.tempStor.rateTemp.X0.7.m qfqm.tempStor.rateTemp.X0.8.m
## 1 0 0 0
## qfqm.tempStor.rateTemp.X0.9.m qfqm.tempStor.rateTemp.X1.m qfqm.tempStor.rateTemp.X1.1.m
## 1 0 0 0
## qfqm.tempStor.rateTemp.X1.2.m qfqm.tempStor.rateTemp.X1.3.m qfqm.tempStor.rateTemp.X1.4.m
## 1 0 0 0
## qfqm.tempStor.rateTemp.X1.5.m qfqm.tempStor.rateTemp.X1.6.m qfqm.tempStor.rateTemp.X1.7.m
## 1 0 0 0
## qfqm.tempStor.rateTemp.X1.8.m qfqm.tempStor.rateTemp.X1.9.m qfqm.tempStor.rateTemp.X2.m
## 1 0 0 0
## qfqm.tempStor.rateTemp.X2.1.m qfqm.tempStor.rateTemp.X2.2.m qfqm.tempStor.rateTemp.X2.3.m
## 1 0 0 0
## qfqm.tempStor.rateTemp.X2.4.m qfqm.tempStor.rateTemp.X2.5.m qfqm.tempStor.rateTemp.X2.6.m
## 1 0 0 0
## qfqm.tempStor.rateTemp.X2.7.m qfqm.tempStor.rateTemp.X2.8.m qfqm.tempStor.rateTemp.X2.9.m
## 1 0 0 0
## qfqm.tempStor.rateTemp.X3.m qfqm.tempStor.rateTemp.X3.1.m qfqm.tempStor.rateTemp.X3.2.m
## 1 0 0 0
## qfqm.tempStor.rateTemp.X3.3.m qfqm.tempStor.rateTemp.X3.4.m qfqm.tempStor.rateTemp.X3.5.m
## 1 0 0 0
## qfqm.tempStor.rateTemp.X3.6.m qfqm.tempStor.rateTemp.X3.7.m qfqm.tempStor.rateTemp.X3.8.m
## 1 0 0 0
## qfqm.tempStor.rateTemp.X3.9.m qfqm.tempStor.rateTemp.X4.m qfqm.tempStor.rateTemp.X4.1.m
## 1 0 0 0
## qfqm.tempStor.rateTemp.X4.2.m qfqm.tempStor.rateTemp.X4.3.m qfqm.tempStor.rateTemp.X4.4.m
## 1 0 0 0
## qfqm.tempStor.rateTemp.X4.5.m qfqm.tempStor.rateTemp.X4.6.m qfqm.tempStor.rateTemp.X4.7.m
## 1 0 0 0
## qfqm.tempStor.rateTemp.X4.8.m qfqm.tempStor.rateTemp.X4.9.m qfqm.tempStor.rateTemp.X5.m
## 1 0 0 0
## qfqm.tempStor.rateTemp.X5.1.m qfqm.tempStor.rateTemp.X5.2.m qfqm.tempStor.rateTemp.X5.3.m
## 1 0 0 0
## qfqm.tempStor.rateTemp.X5.4.m qfqm.tempStor.rateTemp.X5.5.m qfqm.tempStor.rateTemp.X5.6.m
## 1 0 0 0
## qfqm.tempStor.rateTemp.X5.7.m qfqm.tempStor.rateTemp.X5.8.m qfqm.tempStor.rateTemp.X5.9.m
## 1 0 0 0
## qfqm.tempStor.rateTemp.X6.m qfqm.tempStor.rateTemp.X6.1.m qfqm.tempStor.rateTemp.X6.2.m
## 1 0 0 0
## qfqm.tempStor.rateTemp.X6.3.m qfqm.tempStor.rateTemp.X6.4.m qfqm.tempStor.rateTemp.X6.5.m
## 1 0 0 0
## qfqm.tempStor.rateTemp.X6.6.m qfqm.tempStor.rateTemp.X6.7.m qfqm.tempStor.rateTemp.X6.8.m
## 1 0 0 0
## qfqm.tempStor.rateTemp.X6.9.m qfqm.tempStor.rateTemp.X7.m qfqm.tempStor.rateTemp.X7.1.m
## 1 0 0 0
## qfqm.tempStor.rateTemp.X7.2.m qfqm.tempStor.rateTemp.X7.3.m qfqm.tempStor.rateTemp.X7.4.m
## 1 0 0 0
## qfqm.tempStor.rateTemp.X7.5.m qfqm.tempStor.rateTemp.X7.6.m qfqm.tempStor.rateTemp.X7.7.m
## 1 0 0 0
## qfqm.tempStor.rateTemp.X7.8.m qfqm.tempStor.rateTemp.X7.9.m qfqm.tempStor.rateTemp.X8.m
## 1 0 0 0
## qfqm.tempStor.rateTemp.X8.1.m qfqm.tempStor.rateTemp.X8.2.m qfqm.tempStor.rateTemp.X8.3.m
## 1 0 0 0
## qfqm.tempStor.rateTemp.X8.4.m
## 1 0
## [ reached 'max' / getOption("max.print") -- omitted 5 rows ]
6. Un-interpolated vertical profile data (Level 2)
The Level 2 data are interpolated in time but not in space. They contain the rates of change at each of the measurement heights.
Again, they can be extracted from the HDF5 files using stackEddy()
with the same syntax:
prof.l2 <- stackEddy(filepath="~/Downloads/filesToStack00200/",
level="dp02")
head(prof.l2$HARV)
## verticalPosition timeBgn timeEnd data.co2Stor.rateRtioMoleDryCo2.mean
## 1 010 2018-06-01 00:00:00 2018-06-01 00:29:59 NaN
## 2 010 2018-06-01 00:30:00 2018-06-01 00:59:59 0.002666576
## 3 010 2018-06-01 01:00:00 2018-06-01 01:29:59 -0.011224223
## 4 010 2018-06-01 01:30:00 2018-06-01 01:59:59 0.006133056
## 5 010 2018-06-01 02:00:00 2018-06-01 02:29:59 -0.019554655
## 6 010 2018-06-01 02:30:00 2018-06-01 02:59:59 -0.007855632
## data.h2oStor.rateRtioMoleDryH2o.mean data.tempStor.rateTemp.mean qfqm.co2Stor.rateRtioMoleDryCo2.qfFinl
## 1 NaN 2.583333e-05 1
## 2 NaN -2.008056e-04 1
## 3 NaN -1.901111e-04 1
## 4 NaN -7.419444e-05 1
## 5 NaN -1.537083e-04 1
## 6 NaN -1.874861e-04 1
## qfqm.h2oStor.rateRtioMoleDryH2o.qfFinl qfqm.tempStor.rateTemp.qfFinl
## 1 1 0
## 2 1 0
## 3 1 0
## 4 1 0
## 5 1 0
## 6 1 0
Note that here, as in the PAR data, there is a verticalPosition field.
It has the same meaning as in the PAR data, indicating the tower level of
the measurement.
7. Calibrated raw data (Level 1)
Level 1 (dp01) data are calibrated, and aggregated in time, but
otherwise untransformed. Use Level 1 data for raw gas
concentrations and atmospheric stable isotopes.
Using stackEddy() to extract Level 1 data requires additional
inputs. The Level 1 files are too large to simply pull out all the
variables by default, and they include multiple averaging intervals,
which can't be merged. So two additional inputs are needed:
-
avg: The averaging interval to extract -
var: One or more variables to extract
What variables are available, at what averaging intervals? Another
function in the neonUtilities package, getVarsEddy(), returns
a list of HDF5 file contents. It requires only one input, a filepath
to a single NEON HDF5 file:
vars <- getVarsEddy("~/Downloads/filesToStack00200/NEON.D01.HARV.DP4.00200.001.nsae.2018-07.basic.20201020T201317Z.h5")
head(vars)
## site level category system hor ver tmi name otype dclass dim oth
## 5 HARV dp01 data amrs 000 060 01m angNedXaxs H5I_DATASET COMPOUND 43200 <NA>
## 6 HARV dp01 data amrs 000 060 01m angNedYaxs H5I_DATASET COMPOUND 43200 <NA>
## 7 HARV dp01 data amrs 000 060 01m angNedZaxs H5I_DATASET COMPOUND 43200 <NA>
## 9 HARV dp01 data amrs 000 060 30m angNedXaxs H5I_DATASET COMPOUND 1440 <NA>
## 10 HARV dp01 data amrs 000 060 30m angNedYaxs H5I_DATASET COMPOUND 1440 <NA>
## 11 HARV dp01 data amrs 000 060 30m angNedZaxs H5I_DATASET COMPOUND 1440 <NA>
Inputs to var can be any values from the name field in the table
returned by getVarsEddy(). Let's take a look at CO2 and
H2O, 13C in CO2 and 18O in
H2O, at 30-minute aggregation. Let's look at Harvard Forest
for these data, since deeper canopies generally have more interesting
profiles:
iso <- stackEddy(filepath="~/Downloads/filesToStack00200/",
level="dp01", var=c("rtioMoleDryCo2","rtioMoleDryH2o",
"dlta13CCo2","dlta18OH2o"), avg=30)
head(iso$HARV)
## verticalPosition timeBgn timeEnd data.co2Stor.rtioMoleDryCo2.mean
## 1 010 2018-06-01 00:00:00 2018-06-01 00:29:59 509.3375
## 2 010 2018-06-01 00:30:00 2018-06-01 00:59:59 502.2736
## 3 010 2018-06-01 01:00:00 2018-06-01 01:29:59 521.6139
## 4 010 2018-06-01 01:30:00 2018-06-01 01:59:59 469.6317
## 5 010 2018-06-01 02:00:00 2018-06-01 02:29:59 484.7725
## 6 010 2018-06-01 02:30:00 2018-06-01 02:59:59 476.8554
## data.co2Stor.rtioMoleDryCo2.min data.co2Stor.rtioMoleDryCo2.max data.co2Stor.rtioMoleDryCo2.vari
## 1 451.4786 579.3518 845.0795
## 2 463.5470 533.6622 161.3652
## 3 442.8649 563.0518 547.9924
## 4 432.6588 508.7463 396.8379
## 5 436.2842 537.4641 662.9449
## 6 443.7055 515.6598 246.6969
## data.co2Stor.rtioMoleDryCo2.numSamp data.co2Turb.rtioMoleDryCo2.mean data.co2Turb.rtioMoleDryCo2.min
## 1 235 NA NA
## 2 175 NA NA
## 3 235 NA NA
## 4 175 NA NA
## 5 235 NA NA
## 6 175 NA NA
## data.co2Turb.rtioMoleDryCo2.max data.co2Turb.rtioMoleDryCo2.vari data.co2Turb.rtioMoleDryCo2.numSamp
## 1 NA NA NA
## 2 NA NA NA
## 3 NA NA NA
## 4 NA NA NA
## 5 NA NA NA
## 6 NA NA NA
## data.h2oStor.rtioMoleDryH2o.mean data.h2oStor.rtioMoleDryH2o.min data.h2oStor.rtioMoleDryH2o.max
## 1 NaN NaN NaN
## 2 NaN NaN NaN
## 3 NaN NaN NaN
## 4 NaN NaN NaN
## 5 NaN NaN NaN
## 6 NaN NaN NaN
## data.h2oStor.rtioMoleDryH2o.vari data.h2oStor.rtioMoleDryH2o.numSamp data.h2oTurb.rtioMoleDryH2o.mean
## 1 NA 0 NA
## 2 NA 0 NA
## 3 NA 0 NA
## 4 NA 0 NA
## 5 NA 0 NA
## 6 NA 0 NA
## data.h2oTurb.rtioMoleDryH2o.min data.h2oTurb.rtioMoleDryH2o.max data.h2oTurb.rtioMoleDryH2o.vari
## 1 NA NA NA
## 2 NA NA NA
## 3 NA NA NA
## 4 NA NA NA
## 5 NA NA NA
## 6 NA NA NA
## data.h2oTurb.rtioMoleDryH2o.numSamp data.isoCo2.dlta13CCo2.mean data.isoCo2.dlta13CCo2.min
## 1 NA NaN NaN
## 2 NA -11.40646 -14.992
## 3 NA NaN NaN
## 4 NA -10.69318 -14.065
## 5 NA NaN NaN
## 6 NA -11.02814 -13.280
## data.isoCo2.dlta13CCo2.max data.isoCo2.dlta13CCo2.vari data.isoCo2.dlta13CCo2.numSamp
## 1 NaN NA 0
## 2 -8.022 1.9624355 305
## 3 NaN NA 0
## 4 -7.385 1.5766385 304
## 5 NaN NA 0
## 6 -7.966 0.9929341 308
## data.isoCo2.rtioMoleDryCo2.mean data.isoCo2.rtioMoleDryCo2.min data.isoCo2.rtioMoleDryCo2.max
## 1 NaN NaN NaN
## 2 458.3546 415.875 531.066
## 3 NaN NaN NaN
## 4 439.9582 415.777 475.736
## 5 NaN NaN NaN
## 6 446.5563 420.845 468.312
## data.isoCo2.rtioMoleDryCo2.vari data.isoCo2.rtioMoleDryCo2.numSamp data.isoCo2.rtioMoleDryH2o.mean
## 1 NA 0 NaN
## 2 953.2212 306 22.11830
## 3 NA 0 NaN
## 4 404.0365 306 22.38925
## 5 NA 0 NaN
## 6 138.7560 309 22.15731
## data.isoCo2.rtioMoleDryH2o.min data.isoCo2.rtioMoleDryH2o.max data.isoCo2.rtioMoleDryH2o.vari
## 1 NaN NaN NA
## 2 21.85753 22.34854 0.01746926
## 3 NaN NaN NA
## 4 22.09775 22.59945 0.02626762
## 5 NaN NaN NA
## 6 22.06641 22.26493 0.00277579
## data.isoCo2.rtioMoleDryH2o.numSamp data.isoH2o.dlta18OH2o.mean data.isoH2o.dlta18OH2o.min
## 1 0 NaN NaN
## 2 85 -12.24437 -12.901
## 3 0 NaN NaN
## 4 84 -12.04580 -12.787
## 5 0 NaN NaN
## 6 80 -11.81500 -12.375
## data.isoH2o.dlta18OH2o.max data.isoH2o.dlta18OH2o.vari data.isoH2o.dlta18OH2o.numSamp
## 1 NaN NA 0
## 2 -11.569 0.03557313 540
## 3 NaN NA 0
## 4 -11.542 0.03970481 539
## 5 NaN NA 0
## 6 -11.282 0.03498614 540
## data.isoH2o.rtioMoleDryH2o.mean data.isoH2o.rtioMoleDryH2o.min data.isoH2o.rtioMoleDryH2o.max
## 1 NaN NaN NaN
## 2 20.89354 20.36980 21.13160
## 3 NaN NaN NaN
## 4 21.12872 20.74663 21.33272
## 5 NaN NaN NaN
## 6 20.93480 20.63463 21.00702
## data.isoH2o.rtioMoleDryH2o.vari data.isoH2o.rtioMoleDryH2o.numSamp qfqm.co2Stor.rtioMoleDryCo2.qfFinl
## 1 NA 0 1
## 2 0.025376207 540 1
## 3 NA 0 1
## 4 0.017612293 540 1
## 5 NA 0 1
## 6 0.003805751 540 1
## qfqm.co2Turb.rtioMoleDryCo2.qfFinl qfqm.h2oStor.rtioMoleDryH2o.qfFinl qfqm.h2oTurb.rtioMoleDryH2o.qfFinl
## 1 NA 1 NA
## 2 NA 1 NA
## 3 NA 1 NA
## 4 NA 1 NA
## 5 NA 1 NA
## 6 NA 1 NA
## qfqm.isoCo2.dlta13CCo2.qfFinl qfqm.isoCo2.rtioMoleDryCo2.qfFinl qfqm.isoCo2.rtioMoleDryH2o.qfFinl
## 1 1 1 1
## 2 0 0 0
## 3 1 1 1
## 4 0 0 0
## 5 1 1 1
## 6 0 0 0
## qfqm.isoH2o.dlta18OH2o.qfFinl qfqm.isoH2o.rtioMoleDryH2o.qfFinl ucrt.co2Stor.rtioMoleDryCo2.mean
## 1 1 1 10.0248527
## 2 0 0 1.1077243
## 3 1 1 7.5181428
## 4 0 0 8.4017805
## 5 1 1 0.9465824
## 6 0 0 1.3629090
## ucrt.co2Stor.rtioMoleDryCo2.vari ucrt.co2Stor.rtioMoleDryCo2.se ucrt.co2Turb.rtioMoleDryCo2.mean
## 1 170.28091 1.8963340 NA
## 2 34.29589 0.9602536 NA
## 3 151.35746 1.5270503 NA
## 4 93.41077 1.5058703 NA
## 5 14.02753 1.6795958 NA
## 6 8.50861 1.1873064 NA
## ucrt.co2Turb.rtioMoleDryCo2.vari ucrt.co2Turb.rtioMoleDryCo2.se ucrt.h2oStor.rtioMoleDryH2o.mean
## 1 NA NA NA
## 2 NA NA NA
## 3 NA NA NA
## 4 NA NA NA
## 5 NA NA NA
## 6 NA NA NA
## ucrt.h2oStor.rtioMoleDryH2o.vari ucrt.h2oStor.rtioMoleDryH2o.se ucrt.h2oTurb.rtioMoleDryH2o.mean
## 1 NA NA NA
## 2 NA NA NA
## 3 NA NA NA
## 4 NA NA NA
## 5 NA NA NA
## 6 NA NA NA
## ucrt.h2oTurb.rtioMoleDryH2o.vari ucrt.h2oTurb.rtioMoleDryH2o.se ucrt.isoCo2.dlta13CCo2.mean
## 1 NA NA NaN
## 2 NA NA 0.5812574
## 3 NA NA NaN
## 4 NA NA 0.3653442
## 5 NA NA NaN
## 6 NA NA 0.2428672
## ucrt.isoCo2.dlta13CCo2.vari ucrt.isoCo2.dlta13CCo2.se ucrt.isoCo2.rtioMoleDryCo2.mean
## 1 NaN NA NaN
## 2 0.6827844 0.08021356 16.931819
## 3 NaN NA NaN
## 4 0.3761155 0.07201605 10.078698
## 5 NaN NA NaN
## 6 0.1544487 0.05677862 7.140787
## ucrt.isoCo2.rtioMoleDryCo2.vari ucrt.isoCo2.rtioMoleDryCo2.se ucrt.isoCo2.rtioMoleDryH2o.mean
## 1 NaN NA NaN
## 2 614.01630 1.764965 0.08848440
## 3 NaN NA NaN
## 4 196.99445 1.149078 0.08917388
## 5 NaN NA NaN
## 6 55.90843 0.670111 NA
## ucrt.isoCo2.rtioMoleDryH2o.vari ucrt.isoCo2.rtioMoleDryH2o.se ucrt.isoH2o.dlta18OH2o.mean
## 1 NaN NA NaN
## 2 0.01226428 0.014335993 0.02544454
## 3 NaN NA NaN
## 4 0.01542679 0.017683602 0.01373503
## 5 NaN NA NaN
## 6 NA 0.005890447 0.01932110
## ucrt.isoH2o.dlta18OH2o.vari ucrt.isoH2o.dlta18OH2o.se ucrt.isoH2o.rtioMoleDryH2o.mean
## 1 NaN NA NaN
## 2 0.003017400 0.008116413 0.06937514
## 3 NaN NA NaN
## 4 0.002704220 0.008582764 0.08489408
## 5 NaN NA NaN
## 6 0.002095066 0.008049170 0.02813808
## ucrt.isoH2o.rtioMoleDryH2o.vari ucrt.isoH2o.rtioMoleDryH2o.se
## 1 NaN NA
## 2 0.009640249 0.006855142
## 3 NaN NA
## 4 0.008572288 0.005710986
## 5 NaN NA
## 6 0.002551672 0.002654748
Let's plot vertical profiles of CO2 and 13C in CO2 on a single day.
Here we'll use the time stamps in a different way, using grep() to select
all of the records for a single day. And discard the verticalPosition
values that are string values - those are the calibration gases.
iso.d <- iso$HARV[grep("2018-06-25", iso$HARV$timeBgn, fixed=T),]
iso.d <- iso.d[-which(is.na(as.numeric(iso.d$verticalPosition))),]
ggplot is well suited to these types of data, let's use it to plot
the profiles. If you don't have the package yet, use install.packages()
to install it first.
library(ggplot2)
Now we can plot CO2 relative to height on the tower, with separate lines for each time interval.
g <- ggplot(iso.d, aes(y=verticalPosition)) +
geom_path(aes(x=data.co2Stor.rtioMoleDryCo2.mean,
group=timeBgn, col=timeBgn)) +
theme(legend.position="none") +
xlab("CO2") + ylab("Tower level")
g
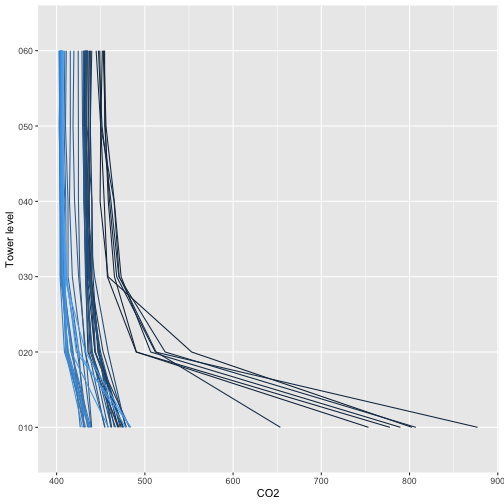
And the same plot for 13C in CO2:
g <- ggplot(iso.d, aes(y=verticalPosition)) +
geom_path(aes(x=data.isoCo2.dlta13CCo2.mean,
group=timeBgn, col=timeBgn)) +
theme(legend.position="none") +
xlab("d13C") + ylab("Tower level")
g
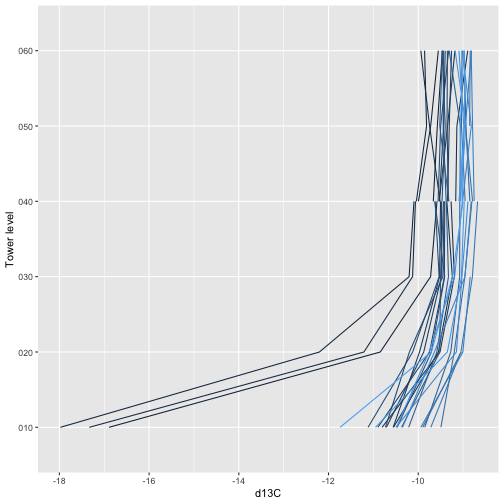
The legends are omitted for space, see if you can use the concentration and isotope ratio buildup and drawdown below the canopy to work out the times of day the different colors represent.