Workshop
Using NEON Data and Teaching Materials with Your Students
BioQuest Summer Workshop: Wicked Problems
Data discovery, manipulation, and analysis are crucial skills for students to be comfortable asking questions using data. As an instructor, you want to be able to use or build teaching materials around data sets that can be used year after year. The standardized data collection and delivery methods for over 180 different data products from the National Ecological Observatory Network (NEON) allow you to use public OERs or build your own materials for your classroom knowing that the same data will be available next year. With this free, standardized data from 81 different NEON field sites from across the US, you can find local or regional data and have students make comparisons across the continent. NEON data can be used at many skill levels; existing OER materials from NEON range from a focus on interpretation of figures to spreadsheet use to reproducible, programming-based data analysis.
In this workshop session, we will spend the first hour exploring open educational resources that use NEON data, with an emphasis on Data Management using NEON’s Small Mammal Data with Accompanying Lesson on Mark Recapture Analysis (McNeil & Jones, 2018). In the second hour, we will use the NEON data portal to directly access NEON data of interest with a discussion of considerations for using the NEON portal and data with students. Participants will leave this session with the tools and comfort level to discover and access data from the NEON data portal and to be aware of and able to use the open educational resources available from NEON.
This session will be presented as a guided navigation through the available resources, therefore, workshop participants should bring a laptop with Excel or Google Sheets (tablets and phones are not ideal) to be able to participate fully in the workshop.
You can also view session information on the BioQuest Summer Workshop page.
During the first hour of the workshop, we will watch a short video on the National Ecological Observatory Network and answer participant's questions on the project. Then, we will focus on open educational resources using NEON data, in particular the Data Management using NEON Small Mammal Data module (below).
NEON: Data to Understand Changing Ecosystems
Teaching Module
Data Management using NEON Small Mammal Data
Authors: Jim McNeil, Megan A. Jones
Teaching Module Overview
Undergraduate STEM students are graduating into professions that require them to manage and work with data at many points of a data management life cycle. Within ecology, students are presented not only with many opportunities to collect data themselves, but increasingly to access and use public data collected by others. This activity introduces the basic concept of data management from the field through to data analysis. The accompanying presentation materials mention the importance of considering long-term data storage and data analysis using public data.
This data set is a subset of small mammal trapping data from the National Ecological Observatory Network (NEON). The accompanying activity introduces students to proper data management practices including how data moves from collection to analysis. Students perform basic spreadsheet tasks to complete a Lincoln-Peterson mark-recapture calculation to estimate population size for a species of small mammal. Pairs of students will work on different sections of the datasets allowing for comparison between seasons or, if instructors download additional data, between sites and years. Data from six months at NEON’s Smithsonian Conservation Biology Institute (SCBI) field site are included in the materials download. Data from other years or locations can be downloaded directly from the NEON data portal to tailor the activity to a specific location or ecological topic.
Learning objectives are achieved through a variety of tactics. By the end of this activity, students will be able to:
- explain the importance of data management in the scientific process. This is accomplished through discussion with faculty/class. Presentation slides are provided to guide this discussion.
- identify elements of a useful, organized data sheet. This is accomplished through inspection of the provided NEON data sheet and how the different data collected are entered into the data collection sheet.
- create a spreadsheet data table for transcription of field collected data. Students will accomplish this by creating and revising a spreadsheet data table and by comparing their version to the provided standardized spreadsheet data table. Faculty are provided with the example Excel workbook and presentation slides to facilitate this learning.
- explain how NEON small mammal data are collected and understand the format that the data are provided. To accomplish this students will read through the provided data collection protocol, view the .csv file containing NEON small mammal data, and view the variables data file to understand what data are contained in the data files. The NEON small mammal data set, protocols, and metadata are included.
- manipulate data in a spreadsheet program (example provided is Microsoft Excel). An example workbook and student handout are provided to show the steps.
- calculate plot-level species population abundance using the Lincoln-Peterson Mark-Recapture model. Presentation slides are provided to introduce the model, and an example workbook and student instructions are provided to show the analysis steps.
- interpret results in plot level species abundance based on temporal variation, geographic locations, or by species. Faculty are provided with notes to help guide this discussion.
This activity was developed while the NEON project was still in construction. There may be future changes to the format of collected and downloaded data. If using data directly from the NEON Data Portal instead of using the data sets accompanying this activity, we recommend testing out the data each year prior to implementing this activity in the classroom.
This module was originally taught starting with a field component where students accompanied NEON technicians during the small mammal trapping. The rest of the activity was implemented in a classroom with students using personal computers. As a field component is not a possibility for most courses, the initial part of the activity has been modified to include optional videos that instructors can use to show how small mammal trapping is conducted. Instructors are also encouraged to bring small mammal traps and small mammal specimens into the classroom when available. Students will need access to computer to complete this activity, however, we recommend they complete the activity in pairs. This could be presented in a computer lab or in a classroom where student have access to personal computers.
Access the Module
This teaching module was originally published as Jim McNeil and Megan A. Jones. April 2018, posting date. Data Management using National Ecological Observatory Network’s (NEON) Small Mammal Data with Accompanying Lesson on Mark Recapture Analysis. Teaching Issues and Experiments in Ecology, Vol. 13: Practice #9 [online]. http://tiee.esa.org/vol/v13/issues/data_sets/mcneil/abstract.html
This teaching module, along with modifications of it by other faculty, can also be found on QUBESHub: Data Management using National Ecological Observatory Network's (NEON) Small Mammal Data with Accompanying Lesson on Mark Recapture Analysis.
Estimated Duration: 50 minutes
Themes: Data Management, Organismal Data
Audience: Undergraduate courses
Class size: Any (adapt the Q&A/Discussion questions to small group discussions or instructor-led introductions as needed)
Technology Needed
- Instructor:
- Internet access: if presenting, live from the website or accessing videos.
- Computer & projector: if presenting, live from the website or accessing videos.
- Optional: audio capabilities for videos
- Student:
- Computer with a spreadsheet program (Excel, Google Sheets)
- Internet access for downloading data sets (or have them pre-downloaded)
Materials Needed
- Instructor:
- Optional: small mammal specimen/small mammal trapping equipment
- Student:
- Handout with instructions & questions
Learning Objectives
This lesson introduces the basic concept of data management from the field through to data analysis while teaching students about a core ecological field data practice of small mammal trapping.
Science Content Objectives
After the lesson, students will be able to:
- Explain a basic process of data management from collection through analysis and archiving
- Calculate population size estimates using the Lincoln-Peterson Method
Data Skills Objectives
After the lesson, students will be able to:
- Create a well layed out spreadsheet
- Describe data management practices for
The Data Sets
The National Ecological Observatory Network is a program sponsored by the National Science Foundation and operated under cooperative agreement by Battelle Memorial Institute. This material is based in part upon work supported by the National Science Foundation through the NEON Program. The associated datasets are posted for educational purposes only. Data for research purposes should be obtained directly from the National Ecological Observatory Network.
Data Citation: National Ecological Observatory Network. 2017. Data Product: NEON.DP1.10072.001. Provisional data downloaded from http://data.neonscience.org. Battelle, Boulder, CO, USA
Instructional Materials
The materials for this lesson are presented as both a single webpage with the primary content and as downloadable materials that can be used in the classroom.
Use of NEON Teaching Modules
All lessons developed for the National Ecological Observatory Network data are open-access (CC-BY) and designed as a resource for the greater community. General feedback on the lesson is welcomed in the comments section on each page. If you develop additional materials related to or supporting this lesson, and are willing to share them, we provide the opportunity for you
to share them with other instructors. Please contact NEON Data Skills or, if familiar with GitHub, submit an issue or pull request with new materials in the NEON Data Skills repo (tutorials are on master branch). All materials will be reviewed by staff prior to inclusion in or linking from the NEON Data Skills materials.
During the second hour of the workshop, we will focus on accessing data from the NEON data portal. We will also preview the neonUtilities R package available from NEONScience on GitHub. Details on how to use the package in the tutorial below.
Use the neonUtilities Package to Access NEON Data
This tutorial provides an overview of functions in the
neonUtilities package in R and the
neonutilities package in Python. These packages provide a
toolbox of basic functionality for working with NEON data.
This tutorial is primarily an index of functions and their inputs;
for more in-depth guidance in using these functions to work with NEON
data, see the
Download
and Explore tutorial. If you are already familiar with the
neonUtilities package, and need a quick reference guide to
function inputs and notation, see the
neonUtilities
cheat sheet.
Function index
The neonUtilities/neonutilities package
contains several functions (use the R and Python tabs to see the syntax
in each language):
R
stackByTable(): Takes zip files downloaded from the Data Portal or downloaded byzipsByProduct(), unzips them, and joins the monthly files by data table to create a single file per table.zipsByProduct(): A wrapper for the NEON API; downloads data based on data product and site criteria. Stores downloaded data in a format that can then be joined bystackByTable().loadByProduct(): Combines the functionality ofzipsByProduct(),
stackByTable(), andreadTableNEON(): Downloads the specified data, stacks the files, and loads the files to the R environment.byFileAOP(): A wrapper for the NEON API; downloads remote sensing data based on data product, site, and year criteria. Preserves the file structure of the original data.byTileAOP(): Downloads remote sensing data for the specified data product, subset to tiles that intersect a list of coordinates.readTableNEON(): Reads NEON data tables into R, using the variables file to assign R classes to each column.getCitation(): Get a BibTeX citation for a particular data product and release.
Python
stack_by_table(): Takes zip files downloaded from the Data Portal or downloaded byzips_by_product(), unzips them, and joins the monthly files by data table to create a single file per table.zips_by_product(): A wrapper for the NEON API; downloads data based on data product and site criteria. Stores downloaded data in a format that can then be joined bystack_by_table().load_by_product(): Combines the functionality ofzips_by_product(),
stack_by_table(), andread_table_neon(): Downloads the specified data, stacks the files, and loads the files to the R environment.by_file_aop(): A wrapper for the NEON API; downloads remote sensing data based on data product, site, and year criteria. Preserves the file structure of the original data.by_tile_aop(): Downloads remote sensing data for the specified data product, subset to tiles that intersect a list of coordinates.read_table_neon(): Reads NEON data tables into R, using the variables file to assign R classes to each column.get_citation(): Get a BibTeX citation for a particular data product and release.
If you are only interested in joining data
files downloaded from the NEON Data Portal, you will only need to use
stackByTable(). Follow the instructions in the first
section of the
Download
and Explore tutorial.
Install and load packages
First, install and load the package. The installation step only needs to be run once, and then periodically to update when new package versions are released. The load step needs to be run every time you run your code.
R
##
## # install neonUtilities - can skip if already installed
## install.packages("neonUtilities")
##
## # load neonUtilities
library(neonUtilities)
## Python
# install neonutilities - can skip if already installed
# do this in the command line
pip install neonutilities
# load neonutilities in working environment
import neonutilities as nuDownload files and load to working environment
The most popular function in neonUtilities is
loadByProduct() (or load_by_product() in
neonutilities). This function downloads data from the NEON
API, merges the site-by-month files, and loads the resulting data tables
into the programming environment, classifying each variable’s data type
appropriately. It combines the actions of the
zipsByProduct(), stackByTable(), and
readTableNEON() functions, described below.
This is a popular choice because it ensures you’re always working with the latest data, and it ends with ready-to-use tables. However, if you use it in a workflow you run repeatedly, keep in mind it will re-download the data every time.
loadByProduct() works on most observational (OS) and
sensor (IS) data, but not on surface-atmosphere exchange (SAE) data,
remote sensing (AOP) data, and some of the data tables in the microbial
data products. For functions that download AOP data, see the
byFileAOP() and byTileAOP() sections in this
tutorial. For functions that work with SAE data, see the
NEON
eddy flux data tutorial. SAE functions are not yet available in
Python.
The inputs to loadByProduct() control which data to
download and how to manage the processing:
R
dpID: The data product ID, e.g. DP1.00002.001site: Defaults to “all”, meaning all sites with available data; can be a vector of 4-letter NEON site codes, e.g.c("HARV","CPER","ABBY").startdateandenddate: Defaults to NA, meaning all dates with available data; or a date in the form YYYY-MM, e.g. 2017-06. Since NEON data are provided in month packages, finer scale querying is not available. Both start and end date are inclusive.package: Either basic or expanded data package. Expanded data packages generally include additional information about data quality, such as chemical standards and quality flags. Not every data product has an expanded package; if the expanded package is requested but there isn’t one, the basic package will be downloaded.timeIndex: Defaults to “all”, to download all data; or the number of minutes in the averaging interval. See example below; only applicable to IS data.release: Specify a particular data Release, e.g."RELEASE-2024". Defaults to the most recent Release. For more details and guidance, see the Release and Provisional tutorial.include.provisional: T or F: Should provisional data be downloaded? Ifreleaseis not specified, set to T to include provisional data in the download. Defaults to F.savepath: the file path you want to download to; defaults to the working directory.check.size: T or F: should the function pause before downloading data and warn you about the size of your download? Defaults to T; if you are using this function within a script or batch process you will want to set it to F.token: Optional API token for faster downloads. See the API token tutorial.nCores: Number of cores to use for parallel processing. Defaults to 1, i.e. no parallelization.
Python
dpid: the data product ID, e.g. DP1.00002.001site: defaults to “all”, meaning all sites with available data; can be a list of 4-letter NEON site codes, e.g.["HARV","CPER","ABBY"].startdateandenddate: defaults to NA, meaning all dates with available data; or a date in the form YYYY-MM, e.g. 2017-06. Since NEON data are provided in month packages, finer scale querying is not available. Both start and end date are inclusive.package: either basic or expanded data package. Expanded data packages generally include additional information about data quality, such as chemical standards and quality flags. Not every data product has an expanded package; if the expanded package is requested but there isn’t one, the basic package will be downloaded.timeindex: defaults to “all”, to download all data; or the number of minutes in the averaging interval. See example below; only applicable to IS data.release: Specify a particular data Release, e.g."RELEASE-2024". Defaults to the most recent Release. For more details and guidance, see the Release and Provisional tutorial.include_provisional: True or False: Should provisional data be downloaded? Ifreleaseis not specified, set to T to include provisional data in the download. Defaults to F.savepath: the file path you want to download to; defaults to the working directory.check_size: True or False: should the function pause before downloading data and warn you about the size of your download? Defaults to True; if you are using this function within a script or batch process you will want to set it to False.token: Optional API token for faster downloads. See the API token tutorial.cloud_mode: Can be set to True if you are working in a cloud environment; provides more efficient data transfer from NEON cloud storage to other cloud environments.progress: Set to False to omit the progress bar during download and stacking.
The dpID (dpid) is the data product
identifier of the data you want to download. The DPID can be found on
the
Explore Data Products page. It will be in the form DP#.#####.###
Demo data download and read
Let’s get triple-aspirated air temperature data (DP1.00003.001) from
Moab and Onaqui (MOAB and ONAQ), from May–August 2018, and name the data
object triptemp:
R
triptemp <- loadByProduct(dpID="DP1.00003.001",
site=c("MOAB","ONAQ"),
startdate="2018-05",
enddate="2018-08")Python
triptemp = nu.load_by_product(dpid="DP1.00003.001",
site=["MOAB","ONAQ"],
startdate="2018-05",
enddate="2018-08")View downloaded data
The object returned by loadByProduct() is a named list
of data tables, or a dictionary of data tables in Python. To work with
each of them, select them from the list.
R
names(triptemp)## [1] "citation_00003_RELEASE-2024" "issueLog_00003"
## [3] "readme_00003" "sensor_positions_00003"
## [5] "TAAT_1min" "TAAT_30min"
## [7] "variables_00003"temp30 <- triptemp$TAAT_30minIf you prefer to extract each table from the list and work with it as
an independent object, you can use the list2env()
function:
list2env(trip.temp, .GlobalEnv)Python
triptemp.keys()## dict_keys(['TAAT_1min', 'TAAT_30min', 'citation_00003_RELEASE-2024', 'issueLog_00003', 'readme_00003', 'sensor_positions_00003', 'variables_00003'])temp30 = triptemp["TAAT_30min"]If you prefer to extract each table from the list and work with it as
an independent object, you can use
globals().update():
globals().update(triptemp)For more details about the contents of the data tables and metadata tables, check out the Download and Explore tutorial.
Join data files: stackByTable()
The function stackByTable() joins the month-by-site
files from a data download. The output will yield data grouped into new
files by table name. For example, the single aspirated air temperature
data product contains 1 minute and 30 minute interval data. The output
from this function is one .csv with 1 minute data and one .csv with 30
minute data.
Depending on your file size this function may run for a while. For example, in testing for this tutorial, 124 MB of temperature data took about 4 minutes to stack. A progress bar will display while the stacking is in progress.
Download the Data
To stack data from the Portal, first download the data of interest
from the NEON
Data Portal. To stack data downloaded from the API, see the
zipsByProduct() section below.
Your data will download from the Portal in a single zipped file.
The stacking function will only work on zipped Comma Separated Value (.csv) files and not the NEON data stored in other formats (HDF5, etc).
Run stackByTable()
The example data below are single-aspirated air temperature.
To run the stackByTable() function, input the file path
to the downloaded and zipped file.
R
# Modify the file path to the file location on your computer
stackByTable(filepath="~neon/data/NEON_temp-air-single.zip")Python
# Modify the file path to the file location on your computer
nu.stack_by_table(filepath="/neon/data/NEON_temp-air-single.zip")In the same directory as the zipped file, you should now have an unzipped directory of the same name. When you open this you will see a new directory called stackedFiles. This directory contains one or more .csv files (depends on the data product you are working with) with all the data from the months & sites you downloaded. There will also be a single copy of the associated variables, validation, and sensor_positions files, if applicable (validation files are only available for observational data products, and sensor position files are only available for instrument data products).
These .csv files are now ready for use with the program of your choice.
To read the data tables, we recommend using
readTableNEON(), which will assign each column to the
appropriate data type, based on the metadata in the variables file. This
ensures time stamps and missing data are interpreted correctly.
Load data to environment
R
SAAT30 <- readTableNEON(
dataFile='~/stackedFiles/SAAT_30min.csv',
varFile='~/stackedFiles/variables_00002.csv'
)Python
SAAT30 = nu.read_table_neon(
dataFile='/stackedFiles/SAAT_30min.csv',
varFile='/stackedFiles/variables_00002.csv'
)Other function inputs
Other input options in stackByTable() are:
savepath: allows you to specify the file path where you want the stacked files to go, overriding the default. Set to"envt"to load the files to the working environment.saveUnzippedFiles: allows you to keep the unzipped, unstacked files from an intermediate stage of the process; by default they are discarded.
Example usage:
R
stackByTable(filepath="~neon/data/NEON_temp-air-single.zip",
savepath="~data/allTemperature", saveUnzippedFiles=T)
tempsing <- stackByTable(filepath="~neon/data/NEON_temp-air-single.zip",
savepath="envt", saveUnzippedFiles=F)
Python
nu.stack_by_table(filepath="/neon/data/NEON_temp-air-single.zip",
savepath="/data/allTemperature",
saveUnzippedFiles=True)
tempsing <- nu.stack_by_table(filepath="/neon/data/NEON_temp-air-single.zip",
savepath="envt",
saveUnzippedFiles=False)
Download files to be stacked: zipsByProduct()
The function zipsByProduct() is a wrapper for the NEON
API, it downloads zip files for the data product specified and stores
them in a format that can then be passed on to
stackByTable().
Input options for zipsByProduct() are the same as those
for loadByProduct() described above.
Here, we’ll download single-aspirated air temperature (DP1.00002.001) data from Wind River Experimental Forest (WREF) for April and May of 2019.
R
zipsByProduct(dpID="DP1.00002.001", site="WREF",
startdate="2019-04", enddate="2019-05",
package="basic", check.size=T)Downloaded files can now be passed to stackByTable() to
be stacked.
stackByTable(filepath=paste(getwd(),
"/filesToStack00002",
sep=""))Python
nu.zips_by_product(dpid="DP1.00002.001", site="WREF",
startdate="2019-04", enddate="2019-05",
package="basic", check_size=True)Downloaded files can now be passed to stackByTable() to
be stacked.
nu.stack_by_table(filepath=os.getcwd()+
"/filesToStack00002")For many sensor data products, download sizes can get very large, and
stackByTable() takes a long time. The 1-minute or 2-minute
files are much larger than the longer averaging intervals, so if you
don’t need high- frequency data, the timeIndex input option
lets you choose which averaging interval to download.
This option is only applicable to sensor (IS) data, since OS data are not averaged.
Download by averaging interval
Download only the 30-minute data for single-aspirated air temperature at WREF:
R
zipsByProduct(dpID="DP1.00002.001", site="WREF",
startdate="2019-04", enddate="2019-05",
package="basic", timeIndex=30,
check.size=T)Python
nu.zips_by_product(dpid="DP1.00002.001", site="WREF",
startdate="2019-04",
enddate="2019-05", package="basic",
timeindex=30, check_size=True)The 30-minute files can be stacked and loaded as usual.
Download remote sensing files
Remote sensing data files can be very large, and NEON remote sensing
(AOP) data are stored in a directory structure that makes them easier to
navigate. byFileAOP() downloads AOP files from the API
while preserving their directory structure. This provides a convenient
way to access AOP data programmatically.
Be aware that downloads from byFileAOP() can take a VERY
long time, depending on the data you request and your connection speed.
You may need to run the function and then leave your machine on and
downloading for an extended period of time.
Here the example download is the Ecosystem Structure data product at Hop Brook (HOPB) in 2017; we use this as the example because it’s a relatively small year-site-product combination.
R
byFileAOP("DP3.30015.001", site="HOPB",
year=2017, check.size=T)Python
nu.by_file_aop(dpid="DP3.30015.001",
site="HOPB", year=2017,
check_size=True)The files should now be downloaded to a new folder in your working directory.
Download remote sensing files for specific coordinates
Often when using remote sensing data, we only want data covering a
certain area - usually the area where we have coordinated ground
sampling. byTileAOP() queries for data tiles containing a
specified list of coordinates. It only works for the tiled, AKA
mosaicked, versions of the remote sensing data, i.e. the ones with data
product IDs beginning with “DP3”.
Here, we’ll download tiles of vegetation indices data (DP3.30026.001) corresponding to select observational sampling plots. For more information about accessing NEON spatial data, see the API tutorial and the in-development geoNEON package.
For now, assume we’ve used the API to look up the plot centroids of plots SOAP_009 and SOAP_011 at the Soaproot Saddle site. You can also look these up in the Spatial Data folder of the document library. The coordinates of the two plots in UTMs are 298755,4101405 and 299296,4101461. These are 40x40m plots, so in looking for tiles that contain the plots, we want to include a 20m buffer. The “buffer” is actually a square, it’s a delta applied equally to both the easting and northing coordinates.
R
byTileAOP(dpID="DP3.30026.001", site="SOAP",
year=2018, easting=c(298755,299296),
northing=c(4101405,4101461),
buffer=20)Python
nu.by_tile_aop(dpid="DP3.30026.001",
site="SOAP", year=2018,
easting=[298755,299296],
northing=[4101405,4101461],
buffer=20)The 2 tiles covering the SOAP_009 and SOAP_011 plots have
been downloaded.
Using neonUtilities in Python
The instructions below will guide you through using the neonUtilities R package in Python, via the rpy2 package. rpy2 creates an R environment you can interact with from Python.
The assumption in this tutorial is that you want to work with NEON data in
Python, but you want to use the handy download and merge functions provided by
the neonUtilities R package to access and format the data for analysis. If
you want to do your analyses in R, use one of the R-based tutorials linked
below.
For more information about the neonUtilities package, and instructions for
running it in R directly, see the Download and Explore tutorial
and/or the neonUtilities tutorial.
Install and set up
Before starting, you will need:
- Python 3 installed. It is probably possible to use this workflow in Python 2, but these instructions were developed and tested using 3.7.4.
- R installed. You don't need to have ever used it directly. We wrote this tutorial using R 4.1.1, but most other recent versions should also work.
-
rpy2installed. Run the line below from the command line, it won't run within a Python script. See Python documentation for more information on how to install packages.rpy2often has install problems on Windows, see "Windows Users" section below if you are running Windows. - You may need to install
pipbefore installingrpy2, if you don't have it installed already.
From the command line, run pip install rpy2
Windows users
The rpy2 package was built for Mac, and doesn't always work smoothly on Windows. If you have trouble with the install, try these steps.
- Add C:\Program Files\R\R-3.3.1\bin\x64 to the Windows Environment Variable “Path”
- Install rpy2 manually from https://www.lfd.uci.edu/~gohlke/pythonlibs/#rpy2
- Pick the correct version. At the download page the portion of the files with cp## relate to the Python version. e.g., rpy2 2.9.2 cp36 cp36m win_amd64.whl is the correct download when 2.9.2 is the latest version of rpy2 and you are running Python 36 and 64 bit Windows (amd64).
- Save the whl file, navigate to it in windows then run pip directly on the file as follows “pip install rpy2 2.9.2 cp36 cp36m win_amd64.whl”
- Add an R_HOME Windows environment variable with the path C:\Program Files\R\R-3.4.3 (or whichever version you are running)
- Add an R_USER Windows environment variable with the path C:\Users\yourUserName\AppData\Local\Continuum\Anaconda3\Lib\site-packages\rpy2
Additional troubleshooting
If you're still having trouble getting R to communicate with Python, you can try pointing Python directly to your R installation path.
- Run
R.home()in R. - Run
import osin Python. - Run
os.environ['R_HOME'] = '/Library/Frameworks/R.framework/Resources'in Python, substituting the file path you found in step 1.
Load packages
Now open up your Python interface of choice (Jupyter notebook, Spyder, etc) and import rpy2 into your session.
import rpy2
import rpy2.robjects as robjects
from rpy2.robjects.packages import importr
Load the base R functionality, using the rpy2 function importr().
base = importr('base')
utils = importr('utils')
stats = importr('stats')
The basic syntax for running R code via rpy2 is package.function(inputs),
where package is the R package in use, function is the name of the function
within the R package, and inputs are the inputs to the function. In other
words, it's very similar to running code in R as package::function(inputs).
For example:
stats.rnorm(6, 0, 1)
FloatVector with 6 elements.
Suppress R warnings. This step can be skipped, but will result in messages getting passed through from R that Python will interpret as warnings.
from rpy2.rinterface_lib.callbacks import logger as rpy2_logger
import logging
rpy2_logger.setLevel(logging.ERROR)
Install the neonUtilities R package. Here I've specified the RStudio
CRAN mirror as the source, but you can use a different one if you
prefer.
You only need to do this step once to use the package, but we update
the neonUtilities package every few months, so reinstalling
periodically is recommended.
This installation step carries out the same steps in the same places on
your hard drive that it would if run in R directly, so if you use R
regularly and have already installed neonUtilities on your machine,
you can skip this step. And be aware, this also means if you install
other packages, or new versions of packages, via rpy2, they'll
be updated the next time you use R, too.
The semicolon at the end of the line (here, and in some other function calls below) can be omitted. It suppresses a note indicating the output of the function is null. The output is null because these functions download or modify files on your local drive, but none of the data are read into the Python or R environments.
utils.install_packages('neonUtilities', repos='https://cran.rstudio.com/');
The downloaded binary packages are in
/var/folders/_k/gbjn452j1h3fk7880d5ppkx1_9xf6m/T//Rtmpl5OpMA/downloaded_packages
Now load the neonUtilities package. This does need to be run every time
you use the code; if you're familiar with R, importr() is roughly
equivalent to the library() function in R.
neonUtilities = importr('neonUtilities')
Join data files: stackByTable()
The function stackByTable() in neonUtilities merges the monthly,
site-level files the NEON Data Portal
provides. Start by downloading the dataset you're interested in from the
Portal. Here, we'll assume you've downloaded IR Biological Temperature.
It will download as a single zip file named NEON_temp-bio.zip. Note the
file path it's saved to and proceed.
Run the stackByTable() function to stack the data. It requires only one
input, the path to the zip file you downloaded from the NEON Data Portal.
Modify the file path in the code below to match the path on your machine.
For additional, optional inputs to stackByTable(), see the R tutorial
for neonUtilities.
neonUtilities.stackByTable(filepath='/Users/Shared/NEON_temp-bio.zip');
Stacking operation across a single core.
Stacking table IRBT_1_minute
Stacking table IRBT_30_minute
Merged the most recent publication of sensor position files for each site and saved to /stackedFiles
Copied the most recent publication of variable definition file to /stackedFiles
Finished: Stacked 2 data tables and 3 metadata tables!
Stacking took 2.019079 secs
All unzipped monthly data folders have been removed.
Check the folder containing the original zip file from the Data Portal;
you should now have a subfolder containing the unzipped and stacked files
called stackedFiles. To import these data to Python, skip ahead to the
"Read downloaded and stacked files into Python" section; to learn how to
use neonUtilities to download data, proceed to the next section.
Download files to be stacked: zipsByProduct()
The function zipsByProduct() uses the NEON API to programmatically download
data files for a given product. The files downloaded by zipsByProduct()
can then be fed into stackByTable().
Run the downloader with these inputs: a data product ID (DPID), a set of 4-letter site IDs (or "all" for all sites), a download package (either basic or expanded), the filepath to download the data to, and an indicator to check the size of your download before proceeding or not (TRUE/FALSE).
The DPID is the data product identifier, and can be found in the data product box on the NEON Explore Data page. Here we'll download Breeding landbird point counts, DP1.10003.001.
There are two differences relative to running zipsByProduct() in R directly:
-
check.sizebecomescheck_size, because dots have programmatic meaning in Python -
TRUE(orT) becomes'TRUE'because the values TRUE and FALSE don't have special meaning in Python the way they do in R, so it interprets them as variables if they're unquoted.
check_size='TRUE' does not work correctly in the Python environment. In R,
it estimates the size of the download and asks you to confirm before
proceeding, and the interactive question and answer don't work correctly
outside R. Set check_size='FALSE' to avoid this problem, but be thoughtful
about the size of your query since it will proceed to download without checking.
neonUtilities.zipsByProduct(dpID='DP1.10003.001',
site=base.c('HARV','BART'),
savepath='/Users/Shared',
package='basic',
check_size='FALSE');
Finding available files
|======================================================================| 100%
Downloading files totaling approximately 4.217543 MB
Downloading 18 files
|======================================================================| 100%
18 files successfully downloaded to /Users/Shared/filesToStack10003
The message output by zipsByProduct() indicates the file path where the
files have been downloaded.
Now take that file path and pass it to stackByTable().
neonUtilities.stackByTable(filepath='/Users/Shared/filesToStack10003');
Unpacking zip files using 1 cores.
Stacking operation across a single core.
Stacking table brd_countdata
Stacking table brd_perpoint
Copied the most recent publication of validation file to /stackedFiles
Copied the most recent publication of categoricalCodes file to /stackedFiles
Copied the most recent publication of variable definition file to /stackedFiles
Finished: Stacked 2 data tables and 4 metadata tables!
Stacking took 0.4586661 secs
All unzipped monthly data folders have been removed.
Read downloaded and stacked files into Python
We've downloaded biological temperature and bird data, and merged the site by month files. Now let's read those data into Python so you can proceed with analyses.
First let's take a look at what's in the output folders.
import os
os.listdir('/Users/Shared/filesToStack10003/stackedFiles/')
['categoricalCodes_10003.csv',
'issueLog_10003.csv',
'brd_countdata.csv',
'brd_perpoint.csv',
'readme_10003.txt',
'variables_10003.csv',
'validation_10003.csv']
os.listdir('/Users/Shared/NEON_temp-bio/stackedFiles/')
['IRBT_1_minute.csv',
'sensor_positions_00005.csv',
'issueLog_00005.csv',
'IRBT_30_minute.csv',
'variables_00005.csv',
'readme_00005.txt']
Each data product folder contains a set of data files and metadata files. Here, we'll read in the data files and take a look at the contents; for more details about the contents of NEON data files and how to interpret them, see the Download and Explore tutorial.
There are a variety of modules and methods for reading tabular data into
Python; here we'll use the pandas module, but feel free to use your own
preferred method.
First, let's read in the two data tables in the bird data:
brd_countdata and brd_perpoint.
import pandas
brd_perpoint = pandas.read_csv('/Users/Shared/filesToStack10003/stackedFiles/brd_perpoint.csv')
brd_countdata = pandas.read_csv('/Users/Shared/filesToStack10003/stackedFiles/brd_countdata.csv')
And take a look at the contents of each file. For descriptions and unit of each
column, see the variables_10003 file.
brd_perpoint
| uid | namedLocation | domainID | siteID | plotID | plotType | pointID | nlcdClass | decimalLatitude | decimalLongitude | ... | endRH | observedHabitat | observedAirTemp | kmPerHourObservedWindSpeed | laboratoryName | samplingProtocolVersion | remarks | measuredBy | publicationDate | release | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 32ab1419-b087-47e1-829d-b1a67a223a01 | BART_025.birdGrid.brd | D01 | BART | BART_025 | distributed | C1 | evergreenForest | 44.060146 | -71.315479 | ... | 56.0 | evergreen forest | 18.0 | 1.0 | Bird Conservancy of the Rockies | NEON.DOC.014041vG | NaN | JRUEB | 20211222T013942Z | RELEASE-2022 |
| 1 | f02e2458-caab-44d8-a21a-b3b210b71006 | BART_025.birdGrid.brd | D01 | BART | BART_025 | distributed | B1 | evergreenForest | 44.060146 | -71.315479 | ... | 56.0 | deciduous forest | 19.0 | 3.0 | Bird Conservancy of the Rockies | NEON.DOC.014041vG | NaN | JRUEB | 20211222T013942Z | RELEASE-2022 |
| 2 | 58ccefb8-7904-4aa6-8447-d6f6590ccdae | BART_025.birdGrid.brd | D01 | BART | BART_025 | distributed | A1 | evergreenForest | 44.060146 | -71.315479 | ... | 56.0 | mixed deciduous/evergreen forest | 17.0 | 0.0 | Bird Conservancy of the Rockies | NEON.DOC.014041vG | NaN | JRUEB | 20211222T013942Z | RELEASE-2022 |
| 3 | 1b14ead4-03fc-4d47-bd00-2f6e31cfe971 | BART_025.birdGrid.brd | D01 | BART | BART_025 | distributed | A2 | evergreenForest | 44.060146 | -71.315479 | ... | 56.0 | deciduous forest | 19.0 | 0.0 | Bird Conservancy of the Rockies | NEON.DOC.014041vG | NaN | JRUEB | 20211222T013942Z | RELEASE-2022 |
| 4 | 3055a0a5-57ae-4e56-9415-eeb7704fab02 | BART_025.birdGrid.brd | D01 | BART | BART_025 | distributed | B2 | evergreenForest | 44.060146 | -71.315479 | ... | 56.0 | deciduous forest | 16.0 | 0.0 | Bird Conservancy of the Rockies | NEON.DOC.014041vG | NaN | JRUEB | 20211222T013942Z | RELEASE-2022 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 1405 | 56d2f3b3-3ee5-41b9-ae22-e78a814d83e4 | HARV_021.birdGrid.brd | D01 | HARV | HARV_021 | distributed | A2 | evergreenForest | 42.451400 | -72.250100 | ... | 71.0 | mixed deciduous/evergreen forest | 16.0 | 1.0 | Bird Conservancy of the Rockies | NEON.DOC.014041vK | NaN | KKLAP | 20221129T224415Z | PROVISIONAL |
| 1406 | 8f61949b-d0cc-49c2-8b59-4e2938286da0 | HARV_021.birdGrid.brd | D01 | HARV | HARV_021 | distributed | A3 | evergreenForest | 42.451400 | -72.250100 | ... | 71.0 | mixed deciduous/evergreen forest | 17.0 | 0.0 | Bird Conservancy of the Rockies | NEON.DOC.014041vK | NaN | KKLAP | 20221129T224415Z | PROVISIONAL |
| 1407 | 36574bab-3725-44d4-b96c-3fc6dcea0765 | HARV_021.birdGrid.brd | D01 | HARV | HARV_021 | distributed | B3 | evergreenForest | 42.451400 | -72.250100 | ... | 71.0 | mixed deciduous/evergreen forest | 19.0 | 0.0 | Bird Conservancy of the Rockies | NEON.DOC.014041vK | NaN | KKLAP | 20221129T224415Z | PROVISIONAL |
| 1408 | eb6dcb4a-cc6c-4ec1-9ee2-6932b7aefc54 | HARV_021.birdGrid.brd | D01 | HARV | HARV_021 | distributed | A1 | evergreenForest | 42.451400 | -72.250100 | ... | 71.0 | deciduous forest | 19.0 | 2.0 | Bird Conservancy of the Rockies | NEON.DOC.014041vK | NaN | KKLAP | 20221129T224415Z | PROVISIONAL |
| 1409 | 51ff3c20-397f-4c88-84e9-f34c2f52d6a8 | HARV_021.birdGrid.brd | D01 | HARV | HARV_021 | distributed | B2 | evergreenForest | 42.451400 | -72.250100 | ... | 71.0 | evergreen forest | 19.0 | 3.0 | Bird Conservancy of the Rockies | NEON.DOC.014041vK | NaN | KKLAP | 20221129T224415Z | PROVISIONAL |
1410 rows × 31 columns
brd_countdata
| uid | namedLocation | domainID | siteID | plotID | plotType | pointID | startDate | eventID | pointCountMinute | ... | vernacularName | observerDistance | detectionMethod | visualConfirmation | sexOrAge | clusterSize | clusterCode | identifiedBy | publicationDate | release | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 4e22256f-5e86-4a2c-99be-dd1c7da7af28 | BART_025.birdGrid.brd | D01 | BART | BART_025 | distributed | C1 | 2015-06-14T09:23Z | BART_025.C1.2015-06-14 | 1 | ... | Black-capped Chickadee | 42.0 | singing | No | Male | 1.0 | NaN | JRUEB | 20211222T013942Z | RELEASE-2022 |
| 1 | 93106c0d-06d8-4816-9892-15c99de03c91 | BART_025.birdGrid.brd | D01 | BART | BART_025 | distributed | C1 | 2015-06-14T09:23Z | BART_025.C1.2015-06-14 | 1 | ... | Red-eyed Vireo | 9.0 | singing | No | Male | 1.0 | NaN | JRUEB | 20211222T013942Z | RELEASE-2022 |
| 2 | 5eb23904-9ae9-45bf-af27-a4fa1efd4e8a | BART_025.birdGrid.brd | D01 | BART | BART_025 | distributed | C1 | 2015-06-14T09:23Z | BART_025.C1.2015-06-14 | 2 | ... | Black-and-white Warbler | 17.0 | singing | No | Male | 1.0 | NaN | JRUEB | 20211222T013942Z | RELEASE-2022 |
| 3 | 99592c6c-4cf7-4de8-9502-b321e925684d | BART_025.birdGrid.brd | D01 | BART | BART_025 | distributed | C1 | 2015-06-14T09:23Z | BART_025.C1.2015-06-14 | 2 | ... | Black-throated Green Warbler | 50.0 | singing | No | Male | 1.0 | NaN | JRUEB | 20211222T013942Z | RELEASE-2022 |
| 4 | 6c07d9fb-8813-452b-8182-3bc5e139d920 | BART_025.birdGrid.brd | D01 | BART | BART_025 | distributed | C1 | 2015-06-14T09:23Z | BART_025.C1.2015-06-14 | 1 | ... | Black-throated Green Warbler | 12.0 | singing | No | Male | 1.0 | NaN | JRUEB | 20211222T013942Z | RELEASE-2022 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 15378 | cffdd5e4-f664-411b-9aea-e6265071332a | HARV_021.birdGrid.brd | D01 | HARV | HARV_021 | distributed | B2 | 2022-06-12T13:31Z | HARV_021.B2.2022-06-12 | 3 | ... | Belted Kingfisher | 37.0 | calling | No | Unknown | 1.0 | NaN | KKLAP | 20221129T224415Z | PROVISIONAL |
| 15379 | 92b58b34-077f-420a-871d-116ac5b1c98a | HARV_021.birdGrid.brd | D01 | HARV | HARV_021 | distributed | B2 | 2022-06-12T13:31Z | HARV_021.B2.2022-06-12 | 5 | ... | Common Yellowthroat | 8.0 | calling | Yes | Male | 1.0 | NaN | KKLAP | 20221129T224415Z | PROVISIONAL |
| 15380 | 06ccb684-da77-4cdf-a8f7-b0d9ac106847 | HARV_021.birdGrid.brd | D01 | HARV | HARV_021 | distributed | B2 | 2022-06-12T13:31Z | HARV_021.B2.2022-06-12 | 1 | ... | Ovenbird | 28.0 | singing | No | Unknown | 1.0 | NaN | KKLAP | 20221129T224415Z | PROVISIONAL |
| 15381 | 0254f165-0052-406e-b9ae-b76ef4109df1 | HARV_021.birdGrid.brd | D01 | HARV | HARV_021 | distributed | B2 | 2022-06-12T13:31Z | HARV_021.B2.2022-06-12 | 2 | ... | Veery | 50.0 | calling | No | Unknown | 1.0 | NaN | KKLAP | 20221129T224415Z | PROVISIONAL |
| 15382 | 432c797d-c4ea-4bfd-901c-5c2481b845c4 | HARV_021.birdGrid.brd | D01 | HARV | HARV_021 | distributed | B2 | 2022-06-12T13:31Z | HARV_021.B2.2022-06-12 | 4 | ... | Pine Warbler | 29.0 | singing | No | Unknown | 1.0 | NaN | KKLAP | 20221129T224415Z | PROVISIONAL |
15383 rows × 24 columns
And now let's do the same with the 30-minute data table for biological temperature.
IRBT30 = pandas.read_csv('/Users/Shared/NEON_temp-bio/stackedFiles/IRBT_30_minute.csv')
IRBT30
| domainID | siteID | horizontalPosition | verticalPosition | startDateTime | endDateTime | bioTempMean | bioTempMinimum | bioTempMaximum | bioTempVariance | bioTempNumPts | bioTempExpUncert | bioTempStdErMean | finalQF | publicationDate | release | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | D18 | BARR | 0 | 10 | 2021-09-01T00:00:00Z | 2021-09-01T00:30:00Z | 7.82 | 7.43 | 8.39 | 0.03 | 1800.0 | 0.60 | 0.00 | 0 | 20211219T025212Z | PROVISIONAL |
| 1 | D18 | BARR | 0 | 10 | 2021-09-01T00:30:00Z | 2021-09-01T01:00:00Z | 7.47 | 7.16 | 7.75 | 0.01 | 1800.0 | 0.60 | 0.00 | 0 | 20211219T025212Z | PROVISIONAL |
| 2 | D18 | BARR | 0 | 10 | 2021-09-01T01:00:00Z | 2021-09-01T01:30:00Z | 7.43 | 6.89 | 8.11 | 0.07 | 1800.0 | 0.60 | 0.01 | 0 | 20211219T025212Z | PROVISIONAL |
| 3 | D18 | BARR | 0 | 10 | 2021-09-01T01:30:00Z | 2021-09-01T02:00:00Z | 7.36 | 6.78 | 8.15 | 0.06 | 1800.0 | 0.60 | 0.01 | 0 | 20211219T025212Z | PROVISIONAL |
| 4 | D18 | BARR | 0 | 10 | 2021-09-01T02:00:00Z | 2021-09-01T02:30:00Z | 6.91 | 6.50 | 7.27 | 0.03 | 1800.0 | 0.60 | 0.00 | 0 | 20211219T025212Z | PROVISIONAL |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 13099 | D18 | BARR | 3 | 0 | 2021-11-30T21:30:00Z | 2021-11-30T22:00:00Z | -14.62 | -14.78 | -14.46 | 0.00 | 1800.0 | 0.57 | 0.00 | 0 | 20211206T221914Z | PROVISIONAL |
| 13100 | D18 | BARR | 3 | 0 | 2021-11-30T22:00:00Z | 2021-11-30T22:30:00Z | -14.59 | -14.72 | -14.50 | 0.00 | 1800.0 | 0.57 | 0.00 | 0 | 20211206T221914Z | PROVISIONAL |
| 13101 | D18 | BARR | 3 | 0 | 2021-11-30T22:30:00Z | 2021-11-30T23:00:00Z | -14.56 | -14.65 | -14.45 | 0.00 | 1800.0 | 0.57 | 0.00 | 0 | 20211206T221914Z | PROVISIONAL |
| 13102 | D18 | BARR | 3 | 0 | 2021-11-30T23:00:00Z | 2021-11-30T23:30:00Z | -14.50 | -14.60 | -14.39 | 0.00 | 1800.0 | 0.57 | 0.00 | 0 | 20211206T221914Z | PROVISIONAL |
| 13103 | D18 | BARR | 3 | 0 | 2021-11-30T23:30:00Z | 2021-12-01T00:00:00Z | -14.45 | -14.57 | -14.32 | 0.00 | 1800.0 | 0.57 | 0.00 | 0 | 20211206T221914Z | PROVISIONAL |
13104 rows × 16 columns
Download remote sensing files: byFileAOP()
The function byFileAOP() uses the NEON API
to programmatically download data files for remote sensing (AOP) data
products. These files cannot be stacked by stackByTable() because they
are not tabular data. The function simply creates a folder in your working
directory and writes the files there. It preserves the folder structure
for the subproducts.
The inputs to byFileAOP() are a data product ID, a site, a year,
a filepath to save to, and an indicator to check the size of the
download before proceeding, or not. As above, set check_size="FALSE"
when working in Python. Be especially cautious about download size
when downloading AOP data, since the files are very large.
Here, we'll download Ecosystem structure (Canopy Height Model) data from Hopbrook (HOPB) in 2017.
neonUtilities.byFileAOP(dpID='DP3.30015.001', site='HOPB',
year='2017', check_size='FALSE',
savepath='/Users/Shared');
Downloading files totaling approximately 147.930656 MB
Downloading 217 files
|======================================================================| 100%
Successfully downloaded 217 files to /Users/Shared/DP3.30015.001
Let's read one tile of data into Python and view it. We'll use the
rasterio and matplotlib modules here, but as with tabular data,
there are other options available.
import rasterio
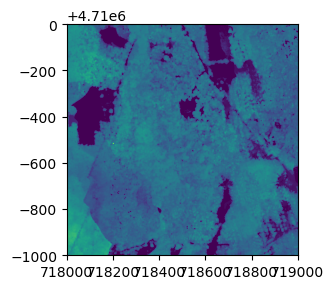
CHMtile = rasterio.open('/Users/Shared/DP3.30015.001/neon-aop-products/2017/FullSite/D01/2017_HOPB_2/L3/DiscreteLidar/CanopyHeightModelGtif/NEON_D01_HOPB_DP3_718000_4709000_CHM.tif')
import matplotlib.pyplot as plt
from rasterio.plot import show
fig, ax = plt.subplots(figsize = (8,3))
show(CHMtile)
<AxesSubplot:>
fig