Tutorial
Random Forest Species Classification using AOP and TOS data in GEE
Authors: John Musinsky, Bridget Hass
Last Updated: Aug 21, 2024
Google Earth Engine has a number of built in machine learning tools that are designed to work with multi-band raster data. This simplifies more complex analyses like classification (eg. classifying land types or species). In this example, we demonstrate species classification using a random forest machine learning model, using NEON AOP reflectance and ecosystem structure (CHM) data, and TOS (Terrestrial Observation System) woody vegetation data to train the model. For this example, we'll use airshed boundary of the site CLBJ (Lyndon B. Johnson National Grassland in north-central Texas).
Objectives
After completing this activity, you will be able to integrate NEON airborne (AOP) and field (TOS) datasets to run supervised classification, which requires:
- Understanding pre-processing steps required for supervised learning classification
- Splitting data into train and test data sets
- Running the Random Forest machine learning model in Google Earth Engine
- Assessing model performance and learn what the different accuracy metrics can tell you
Requirements
- An Earth Engine account. You can sign up for an Earth Engine account here: https://earthengine.google.com/register/
- An understanding of the GEE code editor and the GEE JavaScript API.
- Optionally, complete the first three GEE tutorials in this tutorial series: Intro to AOP Data in Google Earth Engine Tutorial Series
Additional Resources
The links below to the earth engine guides may assist you as you work through this lesson.
Random Forest Machine Learning Workflow
In this tutorial, we will take you through the following steps to classify species at the NEON site CLBJ. Note that these steps closely align with the more general supervised classification steps, described in the Earth Engine Classification Guide .
Workflow Steps:
- Specify the input data for display and use in analysis
- Combine the AOP spectrometer reflectance data with the lidar-derived canopy height model (CHM) to create the predictor dataset
- Create reference (training/test) data for plant species and land cover types based on NEON vegetation structure data
- Fit a random forest model based on spectral/CHM composite to map the distribution of plant species at CLBJ
- Evaluate the model accuracy
Load in and Display the AOP and TOS Data
We'll start by reading in and displaying Map layers of NEON AOP and TOS woody vegetation data. In this first chunk of code, we'll specify the CLBJ location, read in the pre-processed woody vegetation data, as well as the TOS and Airshed boundaries. Details about these boundaries are described on the NEON Flight Box Design webpage.
The plant species data, contained in the CLBJ_veg_2017_filtered feature collection, was derived from the NEON woody plant vegetation structure data product (DP1.10098.001). Land cover classes (grassland, water, shade) were subsequently added to the Feature Collection from visual inspection of the NEON spectrometer reflectance data product (DP3.30006.001).
// Specify CLBJ location
var geo = ee.Geometry.Point([-97.5706464, 33.4045729])
// Load species/land cover samples feature collection to variable (originally a .csv file extracted from NEON woody plant vegetation structure data product (DP1.10098.001))
var CLBJ_veg = ee.FeatureCollection('projects/neon/AOP_NEON_Plots/CLBJ_veg_2017_filtered')
// Load terrestrial observation system (TOS) boundary to a variable
var CLBJ_TOS = ee.FeatureCollection('projects/neon/AOP_TOS_Boundaries/D11_CLBJ_TOS_dissolve')
//Load tower airshed boundary to a variable
var CLBJ_Airshed = ee.FeatureCollection('projects/neon/Airsheds/CLBJ_90percent_footprint')
Next, let's display the Digital Terrain Model (DTM) and Canopy Height Model (CHM) from the 2017 CLBJ collection, masking out the no-data values (-9999).
// Display Digital Terrain Model (DTM) for CLBJ.
// Filter the DEM image collection by the year and geographic location & select the DEM type (DTM)
var clbj_dtm2017 = ee.ImageCollection('projects/neon-prod-earthengine/assets/DEM/001')
.filterDate('2017-01-01', '2017-12-31')
.filterBounds(geo)
.select('DTM')
.first();
// Add the DTM to the map using a histogram stretch based on min and max data values
Map.addLayer(clbj_dtm2017, {min:285, max:1294}, 'CLBJ DTM 2017',0);
// Display Canopy Height Model (CHM) for CLBJ.
var clbj_chm2017 = ee.ImageCollection('projects/neon-prod-earthengine/assets/CHM/001')
.filterDate('2017-01-01', '2017-12-31')
.filterBounds(geo)
.first();
// Add to the map using a histogram stretch based on min and max data values
Map.addLayer(clbj_chm2017, {min:0, max:33}, 'CLBJ CHM 2017',0);
We also want to pull in the Surface Directional Reflectance data . When we do this, we want to keep only the valid bands. Water vapor absorbs light between wavelengths 1340-1445 nm and 1790-1955 nm, and the atmospheric correction that converts radiance to reflectance subsequently results in spikes in reflectance in these two band windows. For more information on the water vapor bands, refer to the lesson Plot Spectral Signatures in Python. We will also remove the last 10 bands, as the bands in this region also tend to be noisy.
To remove bands in GEE, you can specify the bands to exclude (here we named this bandsToRemove) and use the .removeAll function to keep only the valid bands. Note we are including as much spectral information as possible for this tutorial, but you could select a smaller subset of bands and likely obtain similar results. We encourage you to test this out on your own. When running the classification on a larger area, it may be a valuable trade-off to include a smaller number of bands so the code runs faster (or doesn't run out of memory).
// Display Surface Directional Reflectance (HSI_REFL/001) image for CLBJ.
var clbj_sdr2017 = ee.ImageCollection('projects/neon-prod-earthengine/assets/HSI_REFL/001')
.filterDate('2017-01-01', '2017-12-31')
.filterBounds(geo)
.first()
var band_names = clbj_sdr2017.bandNames()
print('Band Names',band_names)
// Remove the "bad" bands - these are the water vapor absorption bands (B195 - B20 and B287 - B310)
// If you look at the data for these bands, you will see these are all set to the same value of -100.
// Also remove the last 10 bands (B416-B425), as those tend to be noisy.
var bands_to_remove = ['B195','B196','B197','B198','B199','B200','B201','B202','B203','B204','B205,',
'B287','B288','B289','B290','B291','B292','B293','B294','B295','B296','B297','B298',
'B299','B300','B301','B302','B303','B304','B305','B306','B307','B308','B309','B310',
'B416','B417','B418','B419','B420','B421','B422','B423','B424','B425']
// Select the inverse of the bad bands to include only the valid bands
var bands_to_keep = band_names.removeAll(bands_to_remove)
var clbj_sdr2017_subset = clbj_sdr2017.select(bands_to_keep)
print('CLBJ SDR 2017 Valid Band Subset')
print(clbj_sdr2017_subset)
// Mask out the no-data values (-9999) in the image and add to the map using a histogram stretch based on lower and upper data values
// var CLBJ_SDR2017mask = CLBJ_SDR2017.updateMask(CLBJ_SDR2017.gte(0.0000)).select(['B053', 'B035', 'B019']);
Map.addLayer(clbj_sdr2017, {min:.5, max:10}, 'CLBJ SDR 2017', 0);
Combine Subset Reflectance and CHM bands
Next we can combine the reflectance bands (381 after we removed the water vapor bands and the last 10 bands) and the CHM band to create a composite multi-band raster that will become our predictor variable (what we use to generate the random forest classification model). We can then crop this to the tower airshed boundary so we can work with a smaller area. This will speed up the process considerably. Optionally, you could classify the full airshed - this code is commented out, if you want to try that instead.
// Combine the reflectance valid data (381 bands) and CHM (1 band) to use in classification
var clbj_sdr_chm2017 = clbj_sdr2017_subset.addBands(clbj_chm2017).aside(print, 'Composite SDR and CHM image');
// Crop the reflectance image/CHM composite to the NEON tower airshed boundary and add to the map
var clbj_sdr_chm_airshed = clbj_sdr_chm2017.clip(clbj_airshed) // comment if uncommenting next line of code
//var CLBJ_SDR2017_airshed = composite // uncomment to classify entire SDR scene
Map.addLayer(clbj_sdr_chm_airshed, {bands:['B053', 'B035', 'B019'], min:.5, max:10}, 'CLBJ-Airshed SDR/CHM 2017');
Display Woody Vegetation Data
The next chunk of code just displays the TOS and Airshed polygon layers, and prints some details about the NEON vegetation structure data to the Console.
// Display the TOS boundary polygon layer
Map.addLayer(clbj_tos.style({width: 3, color: "blue", fillColor: "#00000000"}),{},"CLBJ TOS", 0)
// Display the Airshed polygon layer
Map.addLayer(clbj_airshed.style({width: 3, color: "white", fillColor: "#00000000"}),{},"CLBJ Airshed", 0)
// Print details about the NEON vegetation structure Feature Collection to the Console
// Plant species data were derived from the NEON woody plant vegetation structure data product (DP1.10098.001)
// Land cover classes (grassland, water, shade) were subsequently added to the Feature Collection by visually
// inspecting the NEON spectrometer reflectance data product (DP3.30006.001)
print(clbj_veg, 'All woody plant samples')
If you have run all the code so far, you should be able to see the following layers in the map. Expand the variables printed to the console to make sure the # of bands are correct, and the "bad" (water vapor / noisy) bands are removed.
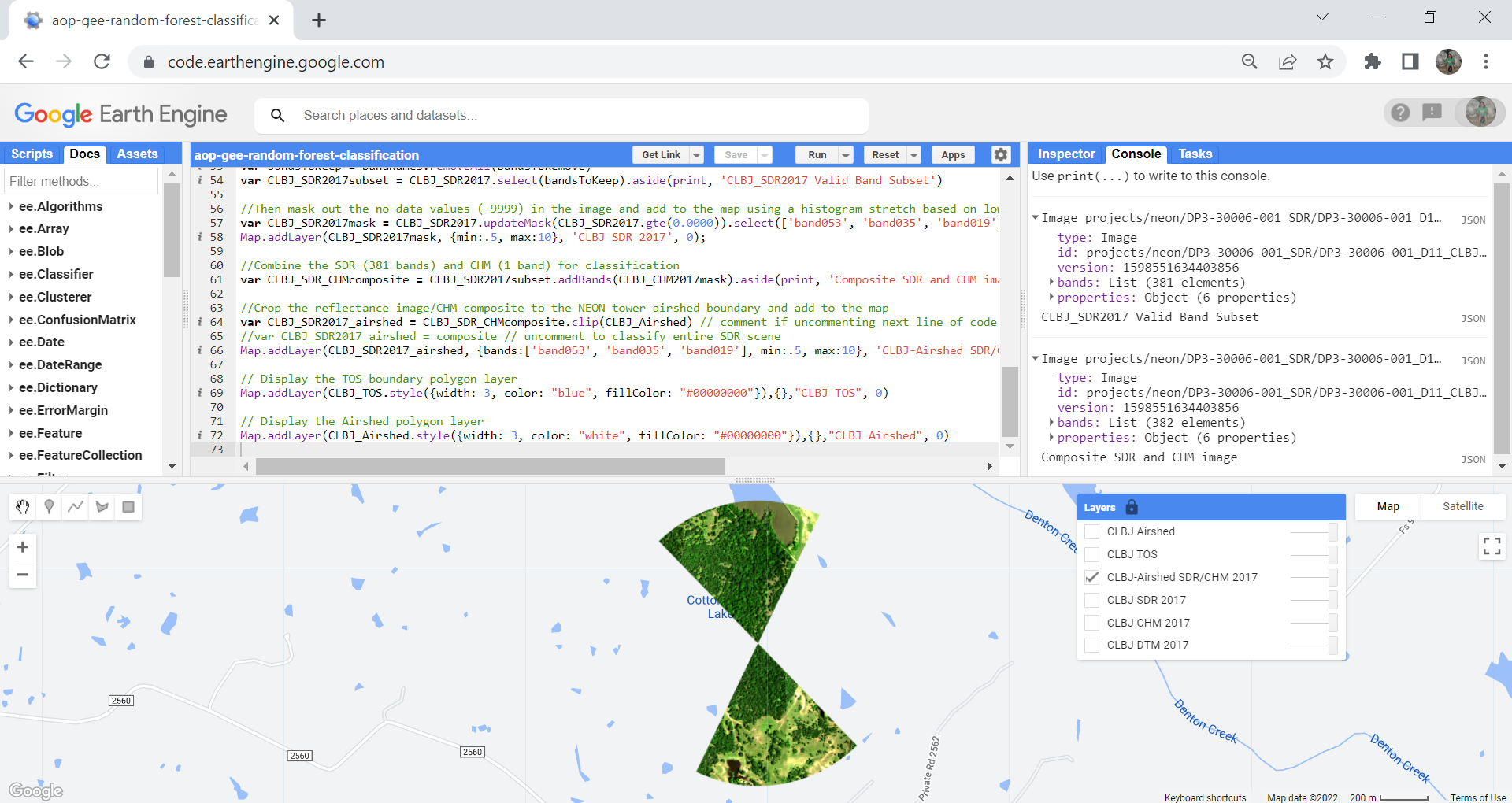
Create Training Data Variables
Now that we've added the relevant AOP data, let's start preparing the training data, which we pulled in at the beginning of the script to the variable CLBJ_veg.
This next chunk of code pulls out each species into separate variables (by their taxon ID), and adds a layer to the Map for each of these variables.
var CLBJ_QUMA3 = CLBJ_veg.filter(ee.Filter.inList('taxonID', ['QUMA3']))
Map.addLayer(CLBJ_QUMA3, {color: 'orange'}, 'Blackjack oak', 0);
var CLBJ_QUST = CLBJ_veg.filter(ee.Filter.inList('taxonID', ['QUST']))
Map.addLayer(CLBJ_QUST, {color: 'darkgreen'}, 'Post oak', 0);
var CLBJ_ULAL = CLBJ_veg.filter(ee.Filter.inList('taxonID', ['ULAL']))
Map.addLayer(CLBJ_ULAL, {color: 'cyan'}, 'Winged elm', 0);
var CLBJ_ULCR = CLBJ_veg.filter(ee.Filter.inList('taxonID', ['ULCR']))
Map.addLayer(CLBJ_ULCR, {color: 'purple'}, 'Cedar elm', 0);
var CLBJ_GRSS = CLBJ_veg.filter(ee.Filter.inList('taxonID', ['GRSS']))
Map.addLayer(CLBJ_GRSS, {color: 'lightgreen'}, 'Grassland', 0);
var CLBJ_WATR = CLBJ_veg.filter(ee.Filter.inList('taxonID', ['WATR']))
Map.addLayer(CLBJ_WATR, {color: 'blue'}, 'Water', 0);
var CLBJ_SHADE = CLBJ_veg.filter(ee.Filter.inList('taxonID', ['SHADE']));
Map.addLayer(CLBJ_SHADE, {color: 'black'}, 'Shade', 0);
Train/Test Split
Once we have the training data for each species, we can split the data for each species into training and test data, using an 80/20 split. The training data will be used later on to train the random forest model, and the test data is used to test the accuracy of the model results on an independent data set.
// Create training and test subsets for each class (i.e., species types) using stratified random sampling (80/20%)
var new_table = CLBJ_CELA.randomColumn({seed: 1});
var CELAtraining = new_table.filter(ee.Filter.lt('random', 0.80));
var CELAtest = new_table.filter(ee.Filter.gte('random', 0.80));
var new_table = CLBJ_JUVI.randomColumn({seed: 1});
var JUVItraining = new_table.filter(ee.Filter.lt('random', 0.80));
var JUVItest = new_table.filter(ee.Filter.gte('random', 0.80));
var new_table = CLBJ_PRME.randomColumn({seed: 1});
var PRMEtraining = new_table.filter(ee.Filter.lt('random', 0.80));
var PRMEtest = new_table.filter(ee.Filter.gte('random', 0.80));
var new_table = CLBJ_QUMA3.randomColumn({seed: 1});
var QUMA3training = new_table.filter(ee.Filter.lt('random', 0.80));
var QUMA3test = new_table.filter(ee.Filter.gte('random', 0.80));
var new_table = CLBJ_QUST.randomColumn({seed: 1});
var QUSTtraining = new_table.filter(ee.Filter.lt('random', 0.80));
var QUSTtest = new_table.filter(ee.Filter.gte('random', 0.80));
var new_table = CLBJ_ULAL.randomColumn({seed: 1});
var ULALtraining = new_table.filter(ee.Filter.lt('random', 0.80));
var ULALtest = new_table.filter(ee.Filter.gte('random', 0.80));
var new_table = CLBJ_ULCR.randomColumn({seed: 1});
var ULCRtraining = new_table.filter(ee.Filter.lt('random', 0.80));
var ULCRtest = new_table.filter(ee.Filter.gte('random', 0.80));
var new_table = CLBJ_GRSS.randomColumn({seed: 1});
var GRSStraining = new_table.filter(ee.Filter.lt('random', 0.80));
var GRSStest = new_table.filter(ee.Filter.gte('random', 0.80));
var new_table = CLBJ_WATR.randomColumn({seed: 1});
var WATRtraining = new_table.filter(ee.Filter.lt('random', 0.80));
var WATRtest = new_table.filter(ee.Filter.gte('random', 0.80));
var new_table = CLBJ_SHADE.randomColumn({seed: 1});
var SHADEtraining = new_table.filter(ee.Filter.lt('random', 0.80));
var SHADEtest = new_table.filter(ee.Filter.gte('random', 0.80));
Now we can merge all the training data for each species together to create the training data (variable), and similarly merge the test data to create the full test data. From those data we'll create a Features variable containing the predictor data (from the spectral and CHM composite) for the training data.
// Combine species-type reference points for training partition
var training = (CELAtraining).merge(JUVItraining).merge(PRMEtraining).merge(QUMA3training).merge(QUSTtraining).merge(ULALtraining).merge(ULCRtraining).merge(GRSStraining).merge(WATRtraining).merge(SHADEtraining).aside(print, 'Training partition');
var test = (CELAtest).merge(JUVItest).merge(PRMEtest).merge(QUMA3test).merge(QUSTtest).merge(ULALtest).merge(ULCRtest).merge(GRSStest).merge(WATRtest).merge(SHADEtest).aside(print, 'Test partition');
var points = training.merge(test).aside(print,'All points');
// Extract spectral signatures from airshed reflectance/CHM image for training sample based on species ID
var Features = CLBJ_SDR2017_airshed.sampleRegions({
collection: training,
properties: ['taxonIDnum'],
scale: 1,
tileScale:16
});
print('Features with spectral signatures:', Features)
Generate and Apply the Random Forest Model
Now that we've assembled the training and test data, and generated predictor data for all of the training data, we can train our random forest model to creat the trainedClassifier variable, and apply it to the reflectance-CHM composite data covering the airshed using .classify(trainedClassifier). Generating the random forest model is pretty simple, once everything is set up!
// Train a 100-tree random forest classifier based on spectral signatures from the training sample for each species
var trainedClassifier = ee.Classifier.smileRandomForest(100)
.train({
features: Features,
classProperty: 'taxonIDnum',
inputProperties: CLBJ_SDR2017_airshed.bandNames()
});
// Classify the reflectance/CHM image from the trained classifier
var classified = CLBJ_SDR2017_airshed.classify(trainedClassifier);
Assess Model Performance
Next we can assess the performance of this classificiation, by looking at some different metrics. The train accuracy should be high (close to 1, or 100%), as it is testing the performance on the data used to generate the model - however, it is not an accurate representation of the actual accuracy. Instead, the test accuracy tends to be a little more reliable, as it is an independent assessment on the separate (test) data set.
// Calulate a confusion matrix and overall accuracy for the training sample
// Note that this overestimates the accuracy of the model since it does not consider the test sample
var trainAccuracy = trainedClassifier.confusionMatrix().accuracy();
print('Train Accuracy', trainAccuracy);
If you look at the console, you should see a train accuracy of 0.973, pretty good! But we expect this to be good, because we are calculating the accuracy of the samples we used to generate the model. It is not representative of the actual model accuracy.
We can also look at some other accuracy metrics. We won't go into details on each of these, but highlight some of the main takeaways for each of the metrics. For more information on accuracy assessments, you can also refer to Qiusheng Wu's Accuracy Assessment for Image Classification Tutorial
- Confusion Matrix: 2D matrix for a classifier based on its training data, where Axis 0 of the matrix corresponds to the input classes (reference data), and axis 1 corresponds to the output classes (classified data). The rows and columns start at class 0 and increase sequentially up to the maximum class value.
- Overall Accuracy: conveys what proportion were mapped correctly out of all of the reference sites (includes training and test data)
- Kappa Coefficient: evaluates how well a classification performs as compared to randomly assigning classes
- Producer's Accuracy: frequency with which a real feature on the ground is correctly shown in the classified map (corresponds to error of omission)
- User's Accuracy: frequency with which a feature in the classified map will actually be present on the ground (corresponds to error of commission)
// Test the classification accuracy (more reliable estimation of accuracy)
// Extract spectral signatures from airshed reflectance image for test sample
var test = CLBJ_SDR2017_airshed.sampleRegions({
collection: test,
properties: ['taxonIDnum'],
scale: 1,
tileScale: 16
});
// Calculate different test accuracy estimates
var Test = test.classify(trainedClassifier);
print('Confusion Matrix', Test.errorMatrix('taxonIDnum', 'classification'));
print('Overall Accuracy', Test.errorMatrix('taxonIDnum', 'classification').accuracy());
print('Kappa Coefficient', Test.errorMatrix('taxonIDnum', 'classification').kappa());
print('Producers Accuracy', Test.errorMatrix('taxonIDnum', 'classification').producersAccuracy());
print('Users Accuracy', Test.errorMatrix('taxonIDnum', 'classification').consumersAccuracy());
When you look in the Console and expand the metrics, you can assess the values. Note each taxon ID is assigned a number so you will need to refer to the order in the code to understand the Confusion Matrix as well as the Producer's and User's Accuracy. Each class is listed below for reference, along with the number of training samples for each of the species, in parentheses. When interpreting accuracy, it's important to consider how many training samples were used to generate the model; the model accuracy is impacted if there is poor representation in the training data.
- CELA - Sugarberry (12)
- JUVI - Eastern Redcedar (35)
- PRME - Mexican Plum (2)
- QUMA3 - Blackjack Oak (26)
- QUST - Post Oak (162)
- ULAL - Winged Elm (5)
- ULCR - Cedar Elm (18)
- GRSS - Grass (8)
- WATR - Water (6)
- SHADE - Shade (9)
Display Model Results
Lastly, we can write a function to display the image classification
// Function used to display training data and image classification
function showTrainingData(){
var colours = ee.List(["yellow", "white", "green", "orange", "darkgreen", "cyan", "purple","lightgreen","blue","black"]);
var lc_type = ee.List(["CELA", "JUVI", "PRME","QUMA3","QUST","ULAL","ULCR","GRSS","WATR","SHADE"]);
var lc_label = ee.List([0, 1, 2, 3, 4, 5, 6, 7, 8, 9]);
var lc_points = ee.FeatureCollection(
lc_type.map(function(lc){
var colour = colours.get(lc_type.indexOf(lc));
return points.filterMetadata("taxonIDnum", "equals", lc_type.indexOf(lc))
.map(function(point){
return point.set('style', {color: colour, pointShape: "diamond", pointSize: 3, width: 2, fillColor: "00000000"});
});
})).flatten();
Map.addLayer(classified, {min: 0, max: 9, palette: ["yellow","white", "green", "orange", "darkgreen", "cyan", "purple", "lightgreen", "blue", "black"]}, 'Classified image', false);
Map.addLayer(lc_points.style({styleProperty: "style"}), {}, 'All training sample points', false);
Map.centerObject(geo, 16)
}
// Display the training data and image classification using the showTrainingData function
showTrainingData();
Here is our final classified image:
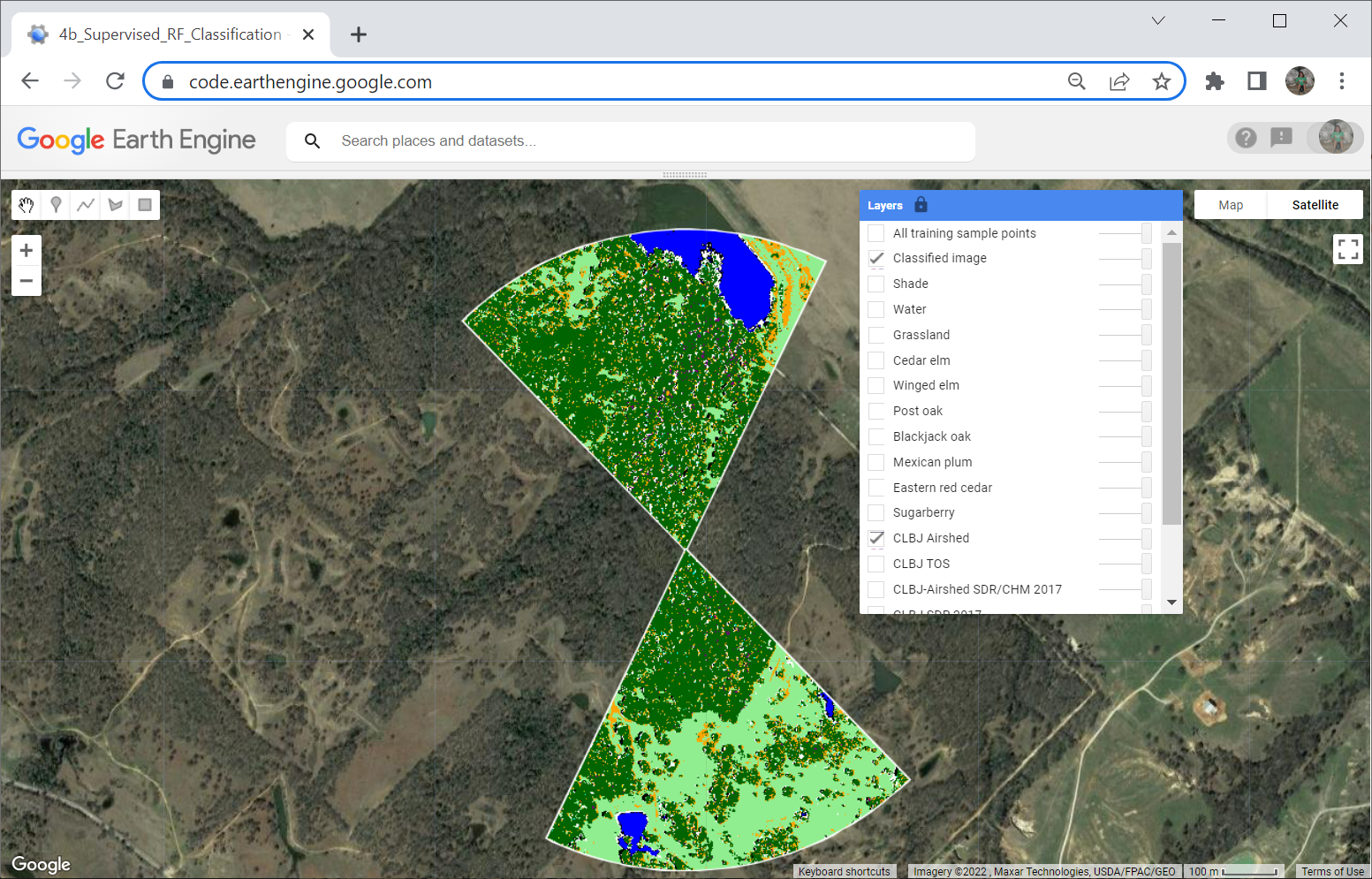
Acknowledgements
This tutorial was modified from a lesson developed by the Organization for Tropical Studies as part of the course Google Earth Engine for Ecology and Conservation. Thank you OTS!