Series
Introduction to Hyperspectral Remote Sensing Data in Python
In this series, we go over the basics of working with the NEON Level 3 NEON Spectrometer orthorectified surface directional reflectance - mosaic data (https://data.neonscience.org/data-products/DP3.30006.001) in Python.
This series covers how to:
- Open hyperspectral reflectance data stored in HDF5 format in Python
- Read in and explore the data and metadata contents
- Use functions in a Python module to efficiently visualize and interact with hyperspectral data cubes
- Extract and plot a single reflectance band and RGB and false-color band combinations
- Read in and plot the spectral signature of a single pixel
- Interactively plot the spectral signature of various pixels within a reflectance tile
Series Objectives
After completing the series you will:
-
Know how to extract data and metadata from HDF5 files in Python
-
Understand how to extract and plot spectra from an HDF5 file
-
Know how to work with groups and datasets within an HDF5 file in Python
-
Create RGB images from different band combinations in a hyperspectral data cube
-
Plot spectral signatures and understand how to remove the water vapor absorption "bad band" windows
-
Create basic vegetation indices, like NDVI, using raster-based calculations
Things You’ll Need To Complete This Series
Setup Python
To complete some of the tutorials in this series, you will need a recent version of Python (3.9+) and, preferably, Jupyter Notebooks installed on your computer. You will also need to install the Python packages requests, gdal, and h5py.
Data
Data are downloaded programmatically in the lessons, but can also be downloaded from the NEON Data Portal. Code for downloading the data are provided in each lesson, but the same dataset is used in multiple lessons, so if you are following in sequence, you do not need to download the data each time.
Read in and visualize hyperspectral data in Python using functions
Authors: Bridget Hass
Last Updated: Jun 25, 2025
In this tutorial, you will learn how to efficiently read in hdf5 reflectance data and metadata, plot a single band and Red-Green-Blue (RGB) band combinations of a reflectance data tile using Python functions created for working with and visualizing NEON AOP hyperspectral data.
This tutorial uses the Level 3 Spectrometer orthorectified surface bidirectional reflectance - mosaic data product.
Learning Objectives
After completing this tutorial, you will be able to:
- Work with custom Python modules and functions for AOP data
- Download and read in tiled NEON AOP reflectance hdf5 data and associated metadata
- Plot a single band of reflectance data
- Stack and plot 3-band combinations to visualize true color and false color images
Things you'll need to complete this tutorial
Python
You will need a current version of Python (3.9+) to complete this tutorial. We also recommend the Jupyter Notebook IDE to follow along with this notebook.
Install Python Packages
- h5py
- gdal
- neonutilities
- pandas
- python-dotenv
- requests
- scikit-image
Data
Data and additional scripts required for this lesson are downloaded programmatically as part of the tutorial. The data used in this lesson were collected over NEON's Disney Wilderness Preserve (DSNY) field site in 2023. The dataset can also be downloaded from the NEON Data Portal.
Other Set-Up Requirements
Set up a NEON user account and token, if you haven't already done so. Follow the tutorial Using an API Token when Accessing NEON Data with neonUtilities to learn how to do this (check the Python tabs in the code cells for the Python syntax).
Note: for this lesson, we have set up the token as an environment variable, following "Option 2: Set token as environment variable" in the linked tutorial above.
Additional Resources
If you are new to AOP hyperspectral data, we recommend exploring the following tutorial series:
Background
We can combine any three bands from the NEON reflectance data to make an RGB image that will depict different information about the Earth's surface. A natural color image, made with bands from the red, green, and blue wavelengths looks close to what we would see with the naked eye. We can also choose band combinations from other wavelenghts, and map them to the red, blue, and green colors to highlight different features. A false color image is made with one or more bands from a non-visible portion of the electromagnetic spectrum that are mapped to red, green, and blue colors. These images can display other information about the landscape that is not easily seen with a natural color image.
The NASA Goddard Media Studio video "Peeling Back Landsat's Layers of Data" gives a good quick overview of natural and false color band combinations. Note that the Landsat multispectral sensor collects information from 11 bands, while NEON AOP hyperspectral data captures information spanning 426 bands!
Peeling Back Landsat's Layers of Data Video
Further Reading
- Check out the NASA Earth Observatory article How to Interpret a False-Color Satellite Image.
- Read the supporting article for the video above, Landsat 8 Onion Skin.
Load Function Module
First, import the required packages and the neon_aop_hyperspectral module, which includes functions that we will use to read in and visualize the hyperspectral hdf5 data.
import dotenv
import h5py
import matplotlib.pyplot as plt
import neonutilities as nu
import numpy as np
import os
import requests
import sys
import time
This next function is a handy way to download the Python module that we will be use in this lesson. This uses the requests package.
# function to download data stored on the internet in a public url to a local file
def download_url(url,download_dir):
if not os.path.isdir(download_dir):
os.makedirs(download_dir)
filename = url.split('/')[-1]
r = requests.get(url, allow_redirects=True)
file_object = open(os.path.join(download_dir,filename),'wb')
file_object.write(r.content)
Download the module from its location on GitHub, add the ../python_modules directory to the path and import the neon_aop_hyperspectral.py module as neon_hs.
module_url = "https://raw.githubusercontent.com/NEONScience/NEON-Data-Skills/main/tutorials/Python/AOP/aop_python_modules/neon_aop_hyperspectral.py"
download_url(module_url,'../python_modules')
# os.listdir('../python_modules') #optionally show the contents of this directory to confirm the file downloaded
sys.path.insert(0, '../python_modules')
# import the neon_aop_hyperspectral module
import neon_aop_hyperspectral as neon_hs
The first function we will use is aop_h5refl2array. We encourage you to look through the code to understand what it is doing behind the scenes. This function automates the steps required to read AOP hdf5 reflectance files into a Python numpy array. This function also cleans the data: it sets any no data values within the reflectance tile to nan (not a number) and applies the reflectance scale factor so the final array that is returned represents unitless scaled reflectance, with values ranging between 0 and 1 (0-100%).
Data Tip: If you forget the inputs to a function or want to see more details on what the function does, you can use help() or ? to display the associated docstrings.
help(neon_hs.aop_h5refl2array)
# neon_hs.aop_h5refl2array? #uncomment for an alternate way to show the help
Help on function aop_h5refl2array in module neon_aop_hyperspectral:
aop_h5refl2array(h5_filename, raster_type_: Literal['Cast_Shadow', 'Data_Selection_Index', 'GLT_Data', 'Haze_Cloud_Water_Map', 'IGM_Data', 'Illumination_Factor', 'OBS_Data', 'Radiance', 'Reflectance', 'Sky_View_Factor', 'to-sensor_Azimuth_Angle', 'to-sensor_Zenith_Angle', 'Visibility_Index_Map', 'Weather_Quality_Indicator'], only_metadata=False)
read in NEON AOP reflectance hdf5 file and return the un-scaled
reflectance array, associated metadata, and wavelengths
Parameters
----------
h5_filename : string
reflectance hdf5 file name, including full or relative path
raster : string
name of raster value to read in; this will typically be the reflectance data,
but other data stored in the h5 file can be accessed as well
valid options:
Cast_Shadow (ATCOR input)
Data_Selection_Index
GLT_Data
Haze_Cloud_Water_Map (ATCOR output)
IGM_Data
Illumination_Factor (ATCOR input)
OBS_Data
Reflectance
Radiance
Sky_View_Factor (ATCOR input)
to-sensor_Azimuth_Angle
to-sensor_Zenith_Angle
Visibility_Index_Map: sea level values of visibility index / total optical thickeness
Weather_Quality_Indicator: estimated percentage of overhead cloud cover during acquisition
Returns
--------
raster_array : ndarray
array of reflectance values
metadata: dictionary
associated metadata containing
bad_band_window1 (tuple)
bad_band_window2 (tuple)
bands: # of bands (float)
data ignore value: value corresponding to no data (float)
epsg: coordinate system code (float)
map info: coordinate system, datum & ellipsoid, pixel dimensions, and origin coordinates (string)
reflectance scale factor: factor by which reflectance is scaled (float)
wavelengths: array
wavelength values, in nm
--------
Example Execution:
--------
refl, refl_metadata = aop_h5refl2array('NEON_D02_SERC_DP3_368000_4306000_reflectance.h5','Reflectance')
Now that we have an idea of how this function works, let's try it out. First, we need to download a reflectance file. We can download a single 1 km x 1 km reflectance data tile for the DSNY site using the neonutilities by_tile_aop function as shown below. This downloads to a data folder specified in savepath. Before downloading a tile, let's take a quick look at when data were collected (and are avaiable) at this site using the list_available_dates function.
# display dates of available data for the directional and bidirectional reflectance data at DNSY
print('Directional reflectance data availability:')
nu.list_available_dates('DP3.30006.001','DSNY') # directional reflectance data ends with .001
print('\nBidirectional reflectance data availability:')
nu.list_available_dates('DP3.30006.002','DSNY') # BRDF and topographic corrected reflectance data ends with .002
Directional reflectance data availability:
RELEASE-2025 Available Dates: 2014-05, 2016-09, 2017-09, 2018-10, 2019-04, 2021-09
Bidirectional reflectance data availability:
PROVISIONAL Available Dates: 2023-04
Next we can also look at the tile extents so we can roughly determine the valid values to enter for the easting and northing, which are input parameters to the by_tile_aop function. First, let's set our NEON token as follows:
dotenv.set_key(dotenv_path=".env", key_to_set="NEON_TOKEN", value_to_set="YOUR_TOKEN_HERE")
dotenv.load_dotenv()
my_token=os.environ.get("NEON_TOKEN")
# optionally display the token to double check
# print('my token: ',my_token)
dsny_bounds = nu.get_aop_tile_extents('DP3.30006.002','DSNY','2023',token=my_token)
Easting Bounds: (451000, 464000)
Northing Bounds: (3099000, 3114000)
# display the first and last UTM coordinates of the DSNY site:
print('First 3 coordinates:\n',dsny_bounds[:3])
print('Last 3 coordinates:\n',dsny_bounds[-3:])
First 3 coordinates:
[(451000, 3103000), (451000, 3104000), (451000, 3105000)]
Last 3 coordinates:
[(463000, 3112000), (464000, 3108000), (464000, 3111000)]
Set up the data directory where we want to download our data.
Data Tip: If you are working from a Windows Operating System (OS), there may be a path length limitation which might cause an error in downloading, since the neon download function maintains the full folder structure the data, as it is stored on Google Cloud Storage (GCS). If you see the following warning: "UserWarning: Filepaths on Windows are limited to 260 characters. Attempting to download a filepath that is 291 characters long. Set the working or savepath directory to be closer to the root directory or enable long path support in Windows.", you will either need to enable long path support in Windows (a quick online search will show you how to do this) or set the savepath directory so that it is shorter. You can use os.path.abspath to see the full path, if you have specified a relative path. For this example, we will set a short savepath by creating a neon_data directly directly under the home directory as follows:
home_dir = os.path.expanduser('~')
data_dir = os.path.join(home_dir,'neon_data')
# optionally display the full path to the data_dir as follows:
# os.path.abspath(data_dir)
nu.by_tile_aop(dpid='DP3.30006.002',
site='DSNY',
year='2023',
easting=454000,
northing=3113000,
include_provisional=True,
savepath=data_dir,
token=my_token)
Provisional NEON data are included. To exclude provisional data, use input parameter include_provisional=False.
Continuing will download 2 NEON data files totaling approximately 713.3 MB. Do you want to proceed? (y/n) y
Define some functions that will help us explore the files that were downloaded. You can also look in the File Explorer (Windows) or Finder (Mac) to check out the contents more interactively.
def list_data_subfolders(data_dir):
"""
Recursively finds and lists subfolders within a directory that contain data (files)
and excludes subfolders that only contain other subfolders.
Args:
data_dir: The path to the root directory to search.
Returns:
A list of paths to the subfolders containing data.
"""
data_subfolders = []
for root, dirs, files in os.walk(data_dir):
# Check if the current directory has both subdirectories and files
if dirs and files:
# Iterate through subdirectories to find those that contain files
for dir_name in dirs:
dir_path = os.path.join(root, dir_name)
if any(os.path.isfile(os.path.join(dir_path, f)) for f in os.listdir(dir_path)):
data_subfolders.append(dir_path)
# If the current directory has no subdirectories, but has files, we still want to keep the directory.
elif files:
if root != data_dir: # Avoid adding the root directory itself if it has files
data_subfolders.append(root)
return data_subfolders
def list_data_files(data_dir):
"""
Lists all files within a specified directory and its subdirectories.
Args:
data_dir (str): The path to the data directory to start the search from.
Returns:
list: A list of full paths to all files found.
"""
all_files = []
for root, _, files in os.walk(data_dir):
for file in files:
full_path = os.path.join(root, file)
all_files.append(full_path)
return all_files
We can use these functions to explore the contents of the data that were downloaded. You can also go into File Explorer (Windows) or Finder (Mac) to explore the contents in a more interactive way.
neon_data_subfolders = list_data_subfolders(data_dir)
# display the paths starting with `neon_data` to shorten:
neon_subfolders_short = [f.replace(home_dir,'') for f in neon_data_subfolders]
neon_subfolders_short
['\\neon_data\\DP3.30006.002\\neon-aop-provisional-products\\2023\\FullSite\\D03\\2023_DSNY_7\\L3\\Spectrometer\\Reflectance',
'\\neon_data\\DP3.30006.002\\neon-publication\\NEON.DOM.SITE.DP3.30006.002\\DSNY\\20230401T000000--20230501T000000\\basic']
Data were downloaded into two nested subfolders. The reflectance data is saved in the path 2023\FullSite\D03\2023_DSNY_7\L3\Spectrometer\Reflectance. This is the standard format where you can expect to find L3 data. Note that before 2023 there is a neon-aop-provisional-products folder. This is because the DSNY data from 2023 is available provisionally. If the data were released, it would be found under neon-aop-products.
Next let's use the list_data_files function to see the actual files that we downloaded. If you included a larger range of points in the Easting and Northing, or used by_file_aop, this list could be much longer.
downloaded_refl_files = list_data_files(data_dir)
# display the files starting with `neon_data` to shorten:
downloaded_refl_files_short = [f.replace(home_dir,'') for f in downloaded_refl_files]
downloaded_refl_files_short
['\\neon_data\\DP3.30006.002\\citation_DP3.30006.002_PROVISIONAL.txt',
'\\neon_data\\DP3.30006.002\\issueLog_DP3.30006.002.csv',
'\\neon_data\\DP3.30006.002\\neon-aop-provisional-products\\2023\\FullSite\\D03\\2023_DSNY_7\\L3\\Spectrometer\\Reflectance\\NEON_D03_DSNY_DP3_454000_3113000_bidirectional_reflectance.h5',
'\\neon_data\\DP3.30006.002\\neon-publication\\NEON.DOM.SITE.DP3.30006.002\\DSNY\\20230401T000000--20230501T000000\\basic\\NEON.D03.DSNY.DP3.30006.002.readme.20241220T001434Z.txt']
# read the h5 reflectance file (including the full path) to the variable h5_file_name
# h5_file_name = data_url.split('/')[-1]
h5_tiles = [f for f in downloaded_refl_files if f.endswith('.h5')]
h5_tile = h5_tiles[0]
# print(f'h5_tile: {h5_tile}')
Now that we've specified our reflectance tile, we can call aop_h5refl2array to read in the reflectance tile as a python array called refl, the metadata into a dictionary called refl_metadata, and the wavelengths into an array. Let's read it it and then take a quick look at the metadata and the first 5 wavelength values.
# read in the reflectance data using the aop_h5refl2array function, this may also take a bit of time
start_time = time.time()
refl, refl_metadata, wavelengths = neon_hs.aop_h5refl2array(h5_tile,'Reflectance')
print("--- It took %s seconds to read in the data ---" % round((time.time() - start_time),0))
Reading in C:\Users\bhass\neon_data\DP3.30006.002\neon-aop-provisional-products\2023\FullSite\D03\2023_DSNY_7\L3\Spectrometer\Reflectance\NEON_D03_DSNY_DP3_454000_3113000_bidirectional_reflectance.h5
--- It took 23.0 seconds to read in the data ---
# display the reflectance metadata dictionary contents
refl_metadata
{'shape': (1000, 1000, 426),
'no_data_value': -9999.0,
'scale_factor': 10000.0,
'bad_band_window1': array([1340, 1445]),
'bad_band_window2': array([1790, 1955]),
'projection': b'+proj=UTM +zone=17 +ellps=WGS84 +datum=WGS84 +units=m +no_defs',
'EPSG': 32617,
'res': {'pixelWidth': 1.0, 'pixelHeight': 1.0},
'extent': (454000.0, 455000.0, 3113000.0, 3114000.0),
'ext_dict': {'xMin': 454000.0,
'xMax': 455000.0,
'yMin': 3113000.0,
'yMax': 3114000.0},
'source': 'C:\\Users\\bhass\\neon_data\\DP3.30006.002\\neon-aop-provisional-products\\2023\\FullSite\\D03\\2023_DSNY_7\\L3\\Spectrometer\\Reflectance\\NEON_D03_DSNY_DP3_454000_3113000_bidirectional_reflectance.h5'}
# display the first 5 values of the wavelengths
wavelengths[:5]
array([383.884003, 388.891693, 393.899506, 398.907196, 403.915009])
We can use the shape method to see the dimensions of the array we read in. Use this method to confirm that the size of the reflectance array makes sense given the hyperspectral data cube, which is 1000 meters x 1000 meters x 426 bands.
refl.shape
(1000, 1000, 426)
Plot a single band
Next we'll use the function plot_aop_refl to plot a single band of the reflectance data. You can use help to understand the required inputs and data types for each of these; only the band and spatial extent are required inputs, the rest are optional inputs. If specified, these optional inputs allow you to set the range color values, specify the axis, add a title, colorbar, colorbar title, and change the colormap (default is to plot in greyscale).
band56 = refl[:,:,55]
neon_hs.plot_aop_refl(band56/refl_metadata['scale_factor'],
refl_metadata['extent'],
colorlimit=(0,0.3),
title='DSNY Tile Band 56',
cmap_title='Reflectance',
colormap='gist_earth')

RGB Plots - Band Stacking
It is often useful to look at several bands together. We can extract and stack three reflectance bands in the red, green, and blue (RGB) spectrums to produce a color image that looks like what we see with our eyes; this is your typical camera image. In the next part of this tutorial, we will learn to stack multiple bands and make a geotif raster from the compilation of these bands. We can see that different combinations of bands allow for different visualizations of the remotely-sensed objects and also conveys useful information about the chemical makeup of the Earth's surface.
We will select bands that fall within the visible range of the electromagnetic spectrum (400-700 nm) and at specific points that correspond to what we see as red, green, and blue.
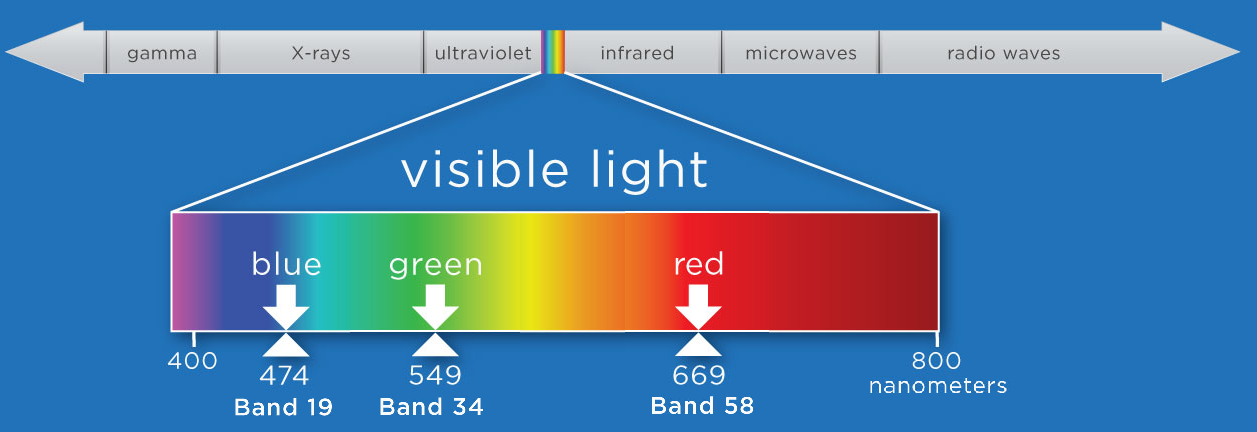
For this exercise, we'll first use the function stack_rgb to extract the bands we want to stack. This function uses splicing to extract the nth band from the reflectance array, and then uses the numpy function stack to create a new 3D array (1000 x 1000 x 3) consisting of only the three bands we want.
# pull out the true-color band combinations
rgb_bands = (58,34,19) # set the red, green, and blue bands
# stack the 3-band combinations (rgb and cir) using stack_rgb function
rgb_unscaled = neon_hs.stack_rgb(refl,rgb_bands)
# apply the reflectance scale factor
rgb = rgb_unscaled/refl_metadata['scale_factor']
We can display the red, green, and blue band center wavelengths, whose indices were defined above. To confirm that these band indices correspond to wavelengths in the expected portion of the spectrum, we can print out the wavelength values in nanometers.
print('Center wavelengths:')
print('Band 58: %.1f' %(wavelengths[57]),'nm')
print('Band 33: %.1f' %(wavelengths[33]),'nm')
print('Band 19: %.1f' %(wavelengths[18]),'nm')
Center wavelengths:
Band 58: 669.3 nm
Band 33: 549.1 nm
Band 19: 474.0 nm
Plot an RGB band combination
Next, we can use the function plot_aop_rgb to plot the band stack as follows:
# plot the true color image (rgb)
neon_hs.plot_aop_rgb(rgb,
refl_metadata['extent'],
plot_title='DSNY Reflectance RGB Image')

False Color Image - Color Infrared (CIR)
We can also create an image from bands outside of the visible spectrum. An image containing one or more bands outside of the visible range is called a false-color image. Here we'll use the green and blue bands as before, but we replace the red band with a near-infrared (NIR) band.
For more information about non-visible wavelengths, false color images, and some frequently used false-color band combinations, refer to NASA's Earth Observatory page.
cir_bands = (90,34,19)
print('Band 90 Center Wavelength = %.1f' %(wavelengths[89]),'nm')
print('Band 34 Center Wavelength = %.1f' %(wavelengths[33]),'nm')
print('Band 19 Center Wavelength = %.1f' %(wavelengths[18]),'nm')
cir = neon_hs.stack_rgb(refl,cir_bands)
neon_hs.plot_aop_rgb(cir,
refl_metadata['extent'],
ls_pct=20,
plot_title='DSNY Color Infrared Image')
Band 90 Center Wavelength = 829.6 nm
Band 34 Center Wavelength = 549.1 nm
Band 19 Center Wavelength = 474.0 nm

Recap
Congratulations! You have successfully downloaded a NEON reflectance tile using the neonutilities by_tile_aop function. You have also pulled in some pre-defined functions and used these to read in and visualize the reflectance data. You are now well poised to start carrying out more in-depth analysis using the hyperspectral data with Python.
References
Kekesi, Alex et al. "NASA | Peeling Back Landsat's Layers of Data". Published on Feb 24, 2014.
Riebeek, Holli. "Why is that Forest Red and that Cloud Blue? How to Interpret a False-Color Satellite Image" . Published on March 4, 2014.
Get Lesson Code
Plot a Spectral Signature from Reflectance Data in Python
Authors: Bridget Hass
Last Updated: Apr 15, 2024
In this tutorial, we will learn how to extract and plot a spectral profile from a single pixel of a reflectance band in a NEON hyperspectral HDF5 file.
This tutorial works with NEON's Level 3 Spectrometer orthorectified surface directional reflectance - mosaic data product.
Objectives
After completing this tutorial, you will be able to:
- Plot the spectral signature of a single pixel
- Remove bad band windows from a spectra
- Use a IPython
widgetto interactively view spectra of various pixels
Install Python Packages
- gdal
- h5py
- requests
- IPython
Data
Data and additional scripts required for this lesson are downloaded programmatically as part of the tutorial.
The hyperspectral imagery file used in this lesson was collected over NEON's Smithsonian Environmental Research Center field site in 2021 and processed at NEON headquarters.
The entire dataset can be accessed on the NEON Data Portal.
In this exercise, we will learn how to extract and plot a spectral profile from a single pixel of a reflectance band in a NEON hyperspectral hdf5 file. To do this, we will use the aop_h5refl2array function to read in and clean our h5 reflectance data, and the Python package pandas to create a dataframe for the reflectance and associated wavelength data.
Spectral Signatures
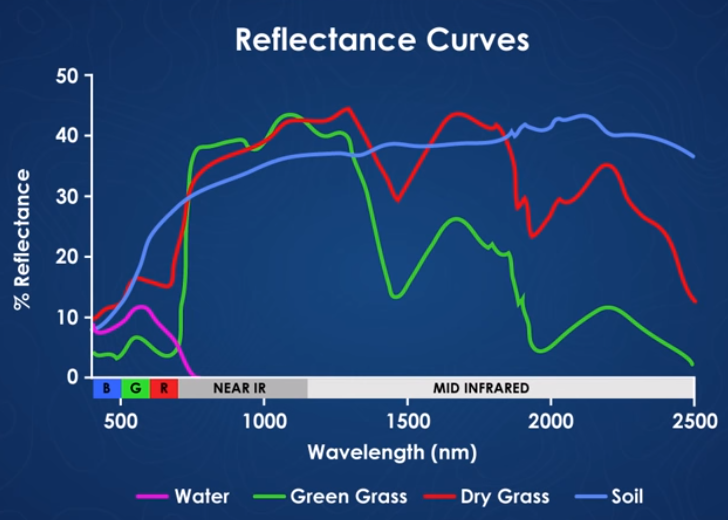
A spectral signature is a plot of the amount of light energy reflected by an object throughout the range of wavelengths in the electromagnetic spectrum. The spectral signature of an object conveys useful information about its structural and chemical composition. We can use these signatures to identify and classify different objects from a spectral image.
For example, vegetation has a distinct spectral signature.
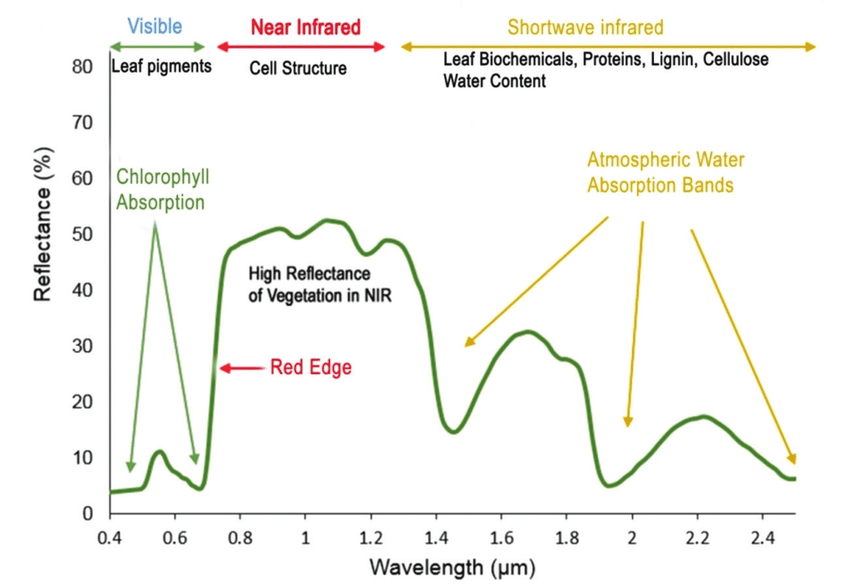
Vegetation has a unique spectral signature characterized by high reflectance in the near infrared wavelengths, and much lower reflectance in the green portion of the visible spectrum. For more details, please see Vegetation Analysis: Using Vegetation Indices in ENVI . We can extract reflectance values in the NIR and visible spectrums from hyperspectral data in order to map vegetation on the earth's surface. You can also use spectral curves as a proxy for vegetation health. We will explore this concept more in the next lesson, where we will caluclate vegetation indices.
import os, sys
import requests
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
This next function is a handy way to download the Python module and data that we will be using for this lesson. This uses the requests package.
# function to download data stored on the internet in a public url to a local file
def download_url(url,download_dir):
if not os.path.isdir(download_dir):
os.makedirs(download_dir)
filename = url.split('/')[-1]
r = requests.get(url, allow_redirects=True)
file_object = open(os.path.join(download_dir,filename),'wb')
file_object.write(r.content)
Download the module from its location on GitHub, add the python_modules to the path and import the neon_aop_hyperspectral.py module.
module_url = "https://raw.githubusercontent.com/NEONScience/NEON-Data-Skills/main/tutorials/Python/AOP/aop_python_modules/neon_aop_hyperspectral.py"
download_url(module_url,'../python_modules')
# os.listdir('../python_modules') #optionally show the contents of this directory to confirm the file downloaded
sys.path.insert(0, '../python_modules')
# import the neon_aop_hyperspectral module, the semicolon supresses an empty plot from displaying
import neon_aop_hyperspectral as neon_hs;
# define the data_url to point to the cloud storage location of the the hyperspectral hdf5 data file
data_url = "https://storage.googleapis.com/neon-aop-products/2021/FullSite/D02/2021_SERC_5/L3/Spectrometer/Reflectance/NEON_D02_SERC_DP3_368000_4306000_reflectance.h5"
# download the h5 data
download_url(data_url,'.\data')
# read the h5 reflectance file (including the full path) to the variable h5_file_name
h5_file_name = data_url.split('/')[-1]
h5_tile = os.path.join(".\data",h5_file_name)
# read in the data using the neon_hs module
serc_refl, serc_refl_md, wavelengths = neon_hs.aop_h5refl2array(h5_tile,'Reflectance')
Reading in .\data\NEON_D02_SERC_DP3_368000_4306000_reflectance.h5
Optionally, you can view the data stored in the metadata dictionary, and print the minimum, maximum, and mean reflectance values in the tile. In order to ignore NaN values, use numpy.nanmin/nanmax/nanmean.
for item in sorted(serc_refl_md):
print(item + ':',serc_refl_md[item])
print('\nSERC Tile Reflectance Stats:')
print('min:',np.nanmin(serc_refl))
print('max:',round(np.nanmax(serc_refl),2))
print('mean:',round(np.nanmean(serc_refl),2))
EPSG: 32618
bad_band_window1: [1340 1445]
bad_band_window2: [1790 1955]
ext_dict: {'xMin': 368000.0, 'xMax': 369000.0, 'yMin': 4306000.0, 'yMax': 4307000.0}
extent: (368000.0, 369000.0, 4306000.0, 4307000.0)
no_data_value: -9999.0
projection: b'+proj=UTM +zone=18 +ellps=WGS84 +datum=WGS84 +units=m +no_defs'
res: {'pixelWidth': 1.0, 'pixelHeight': 1.0}
scale_factor: 10000.0
shape: (1000, 1000, 426)
source: .\data\NEON_D02_SERC_DP3_368000_4306000_reflectance.h5
SERC Tile Reflectance Stats:
min: -100
max: 15459
mean: 1324.72
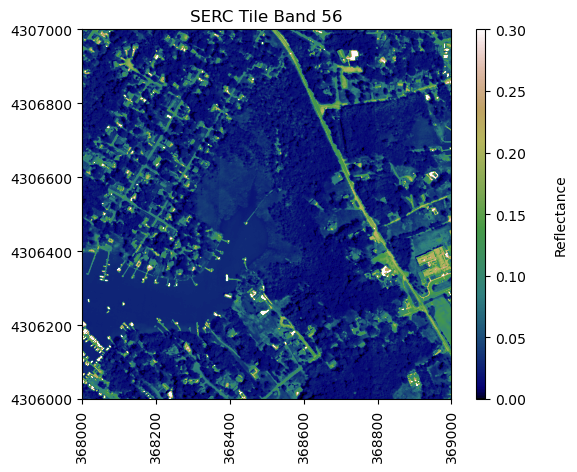
For reference, plot the red band of the tile, using splicing, and the plot_aop_refl function:
sercb56 = serc_refl[:,:,55]/serc_refl_md['scale_factor']
neon_hs.plot_aop_refl(sercb56,
serc_refl_md['extent'],
colorlimit=(0,0.3),
title='SERC Tile Band 56',
cmap_title='Reflectance',
colormap='gist_earth')
We can use pandas to create a dataframe containing the wavelength and reflectance values for a single pixel - in this example, we'll look at the center pixel of the tile (500,500). To extract all reflectance values from a single pixel, use splicing as we did before to select a single band, but now we need to specify (y,x) and select all bands (using :).
serc_pixel_df = pd.DataFrame()
serc_pixel_df['reflectance'] = serc_refl[500,500,:]/serc_refl_md['scale_factor']
serc_pixel_df['wavelengths'] = wavelengths
We can preview the first and last five values of the dataframe using head and tail:
print(serc_pixel_df.head(5))
print(serc_pixel_df.tail(5))
reflectance wavelengths
0 0.0641 383.884003
1 0.0544 388.891693
2 0.0426 393.899506
3 0.0384 398.907196
4 0.0341 403.915009
reflectance wavelengths
421 1.4949 2492.149414
422 1.4948 2497.157227
423 0.6192 2502.165039
424 1.4922 2507.172607
425 1.4922 2512.180420
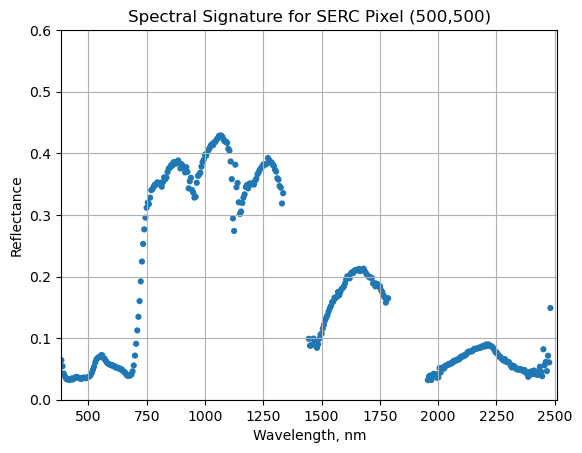
We can now plot the spectra, stored in this dataframe structure. pandas has a built in plotting routine, which can be called by typing .plot at the end of the dataframe.
serc_pixel_df.plot(x='wavelengths',y='reflectance',kind='scatter',edgecolor='none')
plt.title('Spectral Signature for SERC Pixel (500,500)')
ax = plt.gca()
ax.set_xlim([np.min(serc_pixel_df['wavelengths']),np.max(serc_pixel_df['wavelengths'])])
ax.set_ylim(0,0.6)
ax.set_xlabel("Wavelength, nm")
ax.set_ylabel("Reflectance")
ax.grid('on')
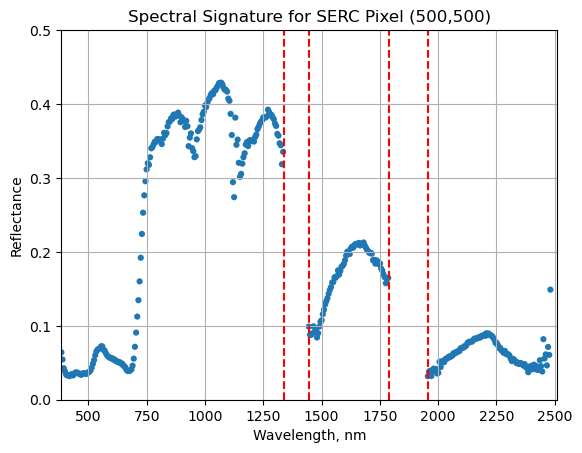
Water Vapor Band Windows
We can see from the spectral profile above that there are spikes in reflectance around ~1400nm and ~1800nm. These result from water vapor which absorbs light between wavelengths 1340-1445 nm and 1790-1955 nm. The atmospheric correction that converts radiance to reflectance subsequently results in a spike at these two bands. The wavelengths of these water vapor bands is stored in the reflectance attributes, which is saved in the reflectance metadata dictionary created with h5refl2array:
bbw1 = serc_refl_md['bad_band_window1'];
bbw2 = serc_refl_md['bad_band_window2'];
print('Bad Band Window 1:',bbw1)
print('Bad Band Window 2:',bbw2)
Bad Band Window 1: [1340 1445]
Bad Band Window 2: [1790 1955]
Below we repeat the plot we made above, but this time draw in the edges of the water vapor band windows that we need to remove.
serc_pixel_df.plot(x='wavelengths',y='reflectance',kind='scatter',edgecolor='none');
plt.title('Spectral Signature for SERC Pixel (500,500)')
ax1 = plt.gca(); ax1.grid('on')
ax1.set_xlim([np.min(serc_pixel_df['wavelengths']),np.max(serc_pixel_df['wavelengths'])]);
ax1.set_ylim(0,0.5)
ax1.set_xlabel("Wavelength, nm"); ax1.set_ylabel("Reflectance")
#Add in red dotted lines to show boundaries of bad band windows:
ax1.plot((1340,1340),(0,1.5), 'r--');
ax1.plot((1445,1445),(0,1.5), 'r--');
ax1.plot((1790,1790),(0,1.5), 'r--');
ax1.plot((1955,1955),(0,1.5), 'r--');
We can now set these bad band windows to nan, along with the last 10 bands, which are also often noisy (as seen in the spectral profile plotted above). First make a copy of the wavelengths so that the original metadata doesn't change.
w = wavelengths.copy() #make a copy to deal with the mutable data type
w[((w >= 1340) & (w <= 1445)) | ((w >= 1790) & (w <= 1955))]=np.nan #can also use bbw1[0] or bbw1[1] to avoid hard-coding in
w[-10:]=np.nan; # the last 10 bands sometimes have noise - best to eliminate
#print(w) #optionally print wavelength values to show that -9999 values are replaced with nan
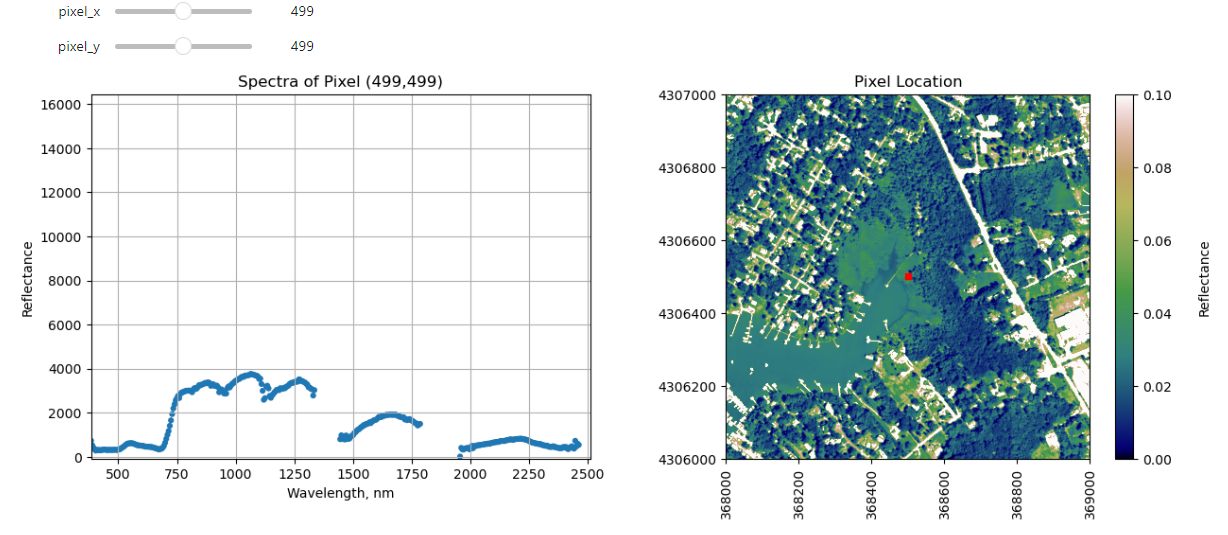
Interactive Spectra Visualization
Finally, we can create a widget to interactively view the spectra of different pixels along the reflectance tile. Run the cell below, and select different pixel_x and pixel_y values to gain a sense of what the spectra look like for different materials on the ground.
#define refl_band, refl, and metadata, as copies of the original serc_refl data
refl_band = sercb56
refl = serc_refl.copy()
metadata = serc_refl_md.copy()
from IPython.html.widgets import *
def interactive_spectra_plot(pixel_x,pixel_y):
reflectance = refl[pixel_y,pixel_x,:]
pixel_df = pd.DataFrame()
pixel_df['reflectance'] = reflectance
pixel_df['wavelengths'] = w
fig = plt.figure(figsize=(15,5))
ax1 = fig.add_subplot(1,2,1)
# fig, axes = plt.subplots(nrows=1, ncols=2)
pixel_df.plot(ax=ax1,x='wavelengths',y='reflectance',kind='scatter',edgecolor='none');
ax1.set_title('Spectra of Pixel (' + str(pixel_x) + ',' + str(pixel_y) + ')')
ax1.set_xlim([np.min(wavelengths),np.max(wavelengths)]);
ax1.set_ylim([np.min(pixel_df['reflectance']),np.max(pixel_df['reflectance']*1.1)])
ax1.set_xlabel("Wavelength, nm"); ax1.set_ylabel("Reflectance")
ax1.grid('on')
ax2 = fig.add_subplot(1,2,2)
plot = plt.imshow(refl_band,extent=metadata['extent'],clim=(0,0.1));
plt.title('Pixel Location');
cbar = plt.colorbar(plot,aspect=20); plt.set_cmap('gist_earth');
cbar.set_label('Reflectance',rotation=90,labelpad=20);
ax2.ticklabel_format(useOffset=False, style='plain') #do not use scientific notation
rotatexlabels = plt.setp(ax2.get_xticklabels(),rotation=90) #rotate x tick labels 90 degrees
ax2.plot(metadata['extent'][0]+pixel_x,metadata['extent'][3]-pixel_y,'s',markersize=5,color='red')
ax2.set_xlim(metadata['extent'][0],metadata['extent'][1])
ax2.set_ylim(metadata['extent'][2],metadata['extent'][3])
interact(interactive_spectra_plot, pixel_x = (0,refl.shape[1]-1,1),pixel_y=(0,refl.shape[0]-1,1));
Get Lesson Code
Calculate NDVI & Extract Spectra Using Masks in Python
Authors: Bridget Hass
Last Updated: Apr 15, 2024
In this tutorial, we will calculate the Normalized Difference Vegetation Index (NDVI) from hyperspectral reflectance data using Python functions.
This tutorial uses the Level 3 Spectrometer orthorectified surface directional reflectance - mosaic.
Objectives
After completing this tutorial, you will be able to:
- Calculate NDVI from hyperspectral data in Python.
Calculate the mean spectr of all pixels whose NDVI is greater than or less than a specified value.I
Install Python Packages
- requests
- pandas
- gdal
- h5py
Data
Data and additional scripts required for this lesson are downloaded programmatically as part of the tutorial.
The hyperspectral imagery file used in this lesson was collected over the National Ecological Observatory Network's Smithsonian Environmental Research Center field site in 2021 and processed at NEON headquarters.
The entire dataset can be accessed on the NEON Data Portal.
Calculate NDVI & Extract Spectra with Masks
Background:
The Normalized Difference Vegetation Index (NDVI) is a standard band-ratio calculation frequently used to analyze ecological remote sensing data. NDVI indicates whether the remotely-sensed target contains live green vegetation. When sunlight strikes objects, certain wavelengths of the electromagnetic spectrum are absorbed and other wavelengths are reflected. The pigment chlorophyll in plant leaves strongly absorbs visible light (with wavelengths in the range of 400-700 nm) for use in photosynthesis. The cell structure of the leaves, however, strongly reflects near-infrared light (wavelengths ranging from 700 - 1100 nm). Plants reflect up to 60% more light in the near infrared portion of the spectrum than they do in the green portion of the spectrum. By calculating the ratio of Near Infrared (NIR) to Visible (VIS) bands in hyperspectral data, we can obtain a metric of vegetation density and health.
The formula for NDVI is: $$NDVI = \frac{(NIR - VIS)}{(NIR+ VIS)}$$
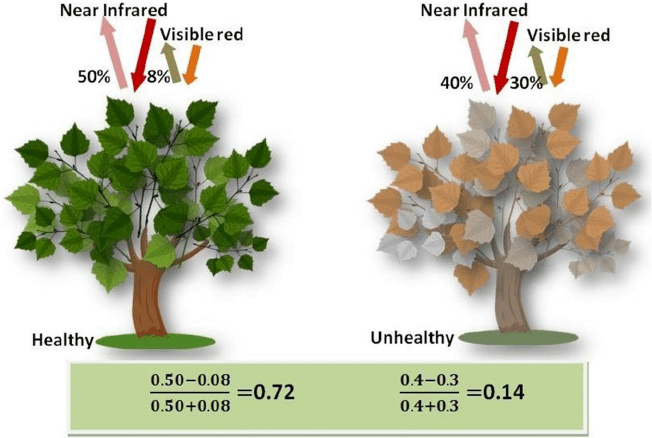
Start by setting plot preferences and loading the neon_aop_hyperspectral.py module:
import os, sys
from copy import copy
import requests
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
This next function is a handy way to download the Python module and data that we will be using for this lesson. This uses the requests package.
# function to download data stored on the internet in a public url to a local file
def download_url(url,download_dir):
if not os.path.isdir(download_dir):
os.makedirs(download_dir)
filename = url.split('/')[-1]
r = requests.get(url, allow_redirects=True)
file_object = open(os.path.join(download_dir,filename),'wb')
file_object.write(r.content)
Download the module from its location on GitHub, add the python_modules to the path and import the neon_aop_hyperspectral.py module as neon_hs.
# download the neon_aop_hyperspectral.py module from GitHub
module_url = "https://raw.githubusercontent.com/NEONScience/NEON-Data-Skills/main/tutorials/Python/AOP/aop_python_modules/neon_aop_hyperspectral.py"
download_url(module_url,'../python_modules')
# add the python_modules to the path and import the python neon download and hyperspectral functions
sys.path.insert(0, '../python_modules')
# import the neon_aop_hyperspectral module, the semicolon supresses an empty plot from displaying
import neon_aop_hyperspectral as neon_hs;
# define the data_url to point to the cloud storage location of the the hyperspectral hdf5 data file
data_url = "https://storage.googleapis.com/neon-aop-products/2021/FullSite/D02/2021_SERC_5/L3/Spectrometer/Reflectance/NEON_D02_SERC_DP3_368000_4306000_reflectance.h5"
# download the h5 data and display how much time it took to download (uncomment 1st and 3rd lines)
# start_time = time.time()
download_url(data_url,'.\data')
# print("--- It took %s seconds to download the data ---" % round((time.time() - start_time),1))
Read in SERC Reflectance Tile
# read the h5 reflectance file (including the full path) to the variable h5_file_name
h5_file_name = data_url.split('/')[-1]
h5_tile = os.path.join(".\data",h5_file_name)
print(f'h5_tile: {h5_tile}')
h5_tile: .\data\NEON_D02_SERC_DP3_368000_4306000_reflectance.h5
# Note you will need to update this filepath for your local machine
serc_refl, serc_refl_md, wavelengths = neon_hs.aop_h5refl2array(h5_tile,'Reflectance')
Reading in .\data\NEON_D02_SERC_DP3_368000_4306000_reflectance.h5
Extract NIR and VIS bands
Now that we have uploaded all the required functions, we can calculate NDVI and plot it. Below we print the center wavelengths that these bands correspond to:
print('band 58 center wavelength (nm): ', wavelengths[57])
print('band 90 center wavelength (nm) : ', wavelengths[89])
band 58 center wavelength (nm): 669.3261
band 90 center wavelength (nm) : 829.5743
Calculate & Plot NDVI
Here we see that band 58 represents red visible light, while band 90 is in the NIR portion of the spectrum. Let's extract these two bands from the reflectance array and calculate the ratio using the numpy.true_divide which divides arrays element-wise. This also handles a case where the denominator = 0, which would otherwise throw a warning or error.
vis = serc_refl[:,:,57]
nir = serc_refl[:,:,89]
# handle a divide by zero by setting the numpy errstate as follows
with np.errstate(divide='ignore', invalid='ignore'):
ndvi = np.true_divide((nir-vis),(nir+vis))
ndvi[ndvi == np.inf] = 0
ndvi = np.nan_to_num(ndvi)
Let's take a look at the min, mean, and max values of NDVI that we calculated:
print(f'NDVI Min: {ndvi.min()}')
print(f'NDVI Mean: {round(ndvi.mean(),2)}')
print(f'NDVI Max: {ndvi.max()}')
NDVI Min: -1.0
NDVI Mean: 0.63
NDVI Max: 1.0
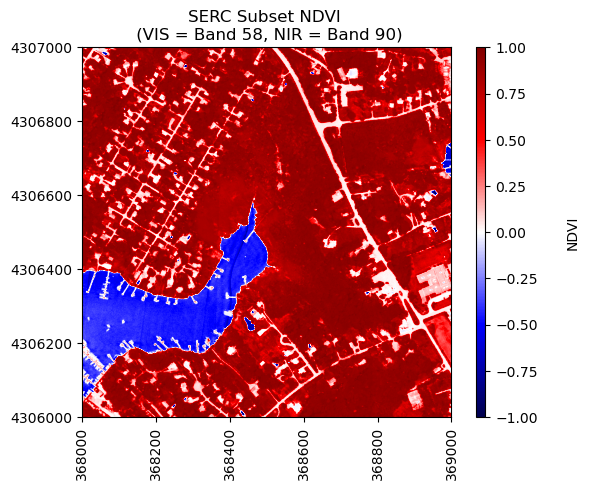
We can use the function plot_aop_refl to plot this, and choose the seismic color pallette to highlight the difference between positive and negative NDVI values. Since this is a normalized index, the values should range from -1 to +1.
neon_hs.plot_aop_refl(ndvi,serc_refl_md['extent'],
colorlimit = (np.min(ndvi),np.max(ndvi)),
title='SERC Subset NDVI \n (VIS = Band 58, NIR = Band 90)',
cmap_title='NDVI',
colormap='seismic')
Extract Spectra Using Masks
In the second part of this tutorial, we will learn how to extract the average spectra of pixels whose NDVI exceeds a specified threshold value. There are several ways to do this using numpy, including the mask functions numpy.ma, as well as numpy.where and finally using boolean indexing.
To start, lets copy the NDVI calculated above and use booleans to create an array only containing NDVI > 0.6.
# make a copy of ndvi
ndvi_gtpt6 = ndvi.copy()
#set all pixels with NDVI < 0.6 to nan, keeping only values > 0.6
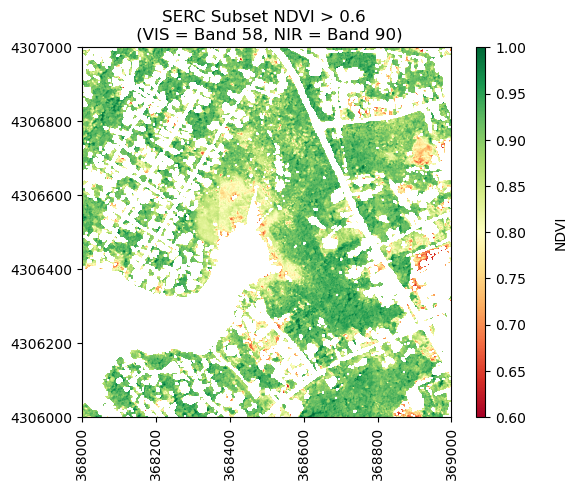
ndvi_gtpt6[ndvi<0.6] = np.nan
print('Mean NDVI > 0.6:',round(np.nanmean(ndvi_gtpt6),2))
Mean NDVI > 0.6: 0.87
Now let's plot the values of NDVI after masking out values < 0.6.
neon_hs.plot_aop_refl(ndvi_gtpt6,
serc_refl_md['extent'],
colorlimit=(0.6,1),
title='SERC Subset NDVI > 0.6 \n (VIS = Band 58, NIR = Band 90)',
cmap_title='NDVI',
colormap='RdYlGn')
Calculate the mean spectra, thresholded by NDVI
Below we will demonstrate how to calculate statistics on arrays where you have applied a mask numpy.ma. In this example, the function calculates the mean spectra for values that remain after masking out values by a specified threshold.
import numpy.ma as ma
def calculate_mean_masked_spectra(refl_array,ndvi,ndvi_threshold,ineq='>'):
mean_masked_refl = np.zeros(refl_array.shape[2])
for i in np.arange(refl_array.shape[2]):
refl_band = refl_array[:,:,i]
if ineq == '>':
ndvi_mask = ma.masked_where((ndvi<=ndvi_threshold) | (np.isnan(ndvi)),ndvi)
elif ineq == '<':
ndvi_mask = ma.masked_where((ndvi>=ndvi_threshold) | (np.isnan(ndvi)),ndvi)
else:
print('ERROR: Invalid inequality. Enter < or >')
masked_refl = ma.MaskedArray(refl_band,mask=ndvi_mask.mask)
mean_masked_refl[i] = ma.mean(masked_refl)
return mean_masked_refl
We can test out this function for various NDVI thresholds. We'll test two together, and you can try out different values on your own. Let's look at the average spectra for healthy vegetation (NDVI > 0.6), and for a lower threshold (NDVI < 0.3).
serc_ndvi_gtpt6 = calculate_mean_masked_spectra(serc_refl,ndvi,0.6)
serc_ndvi_ltpt3 = calculate_mean_masked_spectra(serc_refl,ndvi,0.3,ineq='<')
Finally, we can create a pandas dataframe to plot the mean spectra.
#Remove water vapor bad band windows & last 10 bands
w = wavelengths.copy()
w[((w >= 1340) & (w <= 1445)) | ((w >= 1790) & (w <= 1955))]=np.nan
w[-10:]=np.nan;
nan_ind = np.argwhere(np.isnan(w))
serc_ndvi_gtpt6[nan_ind] = np.nan
serc_ndvi_ltpt3[nan_ind] = np.nan
#Create dataframe with masked NDVI mean spectra, scale by the reflectance scale factor
serc_ndvi_df = pd.DataFrame()
serc_ndvi_df['wavelength'] = w
serc_ndvi_df['mean_refl_ndvi_gtpt6'] = serc_ndvi_gtpt6/serc_refl_md['scale_factor']
serc_ndvi_df['mean_refl_ndvi_ltpt3'] = serc_ndvi_ltpt3/serc_refl_md['scale_factor']
Let's take a look at the first 5 values of this new dataframe:
serc_ndvi_df.head()
| wavelength | mean_refl_ndvi_gtpt6 | mean_refl_ndvi_ltpt3 | |
|---|---|---|---|
| 0 | 383.884003 | 0.055741 | 0.119835 |
| 1 | 388.891693 | 0.036432 | 0.090972 |
| 2 | 393.899506 | 0.027002 | 0.076867 |
| 3 | 398.907196 | 0.022841 | 0.072207 |
| 4 | 403.915009 | 0.018748 | 0.065984 |
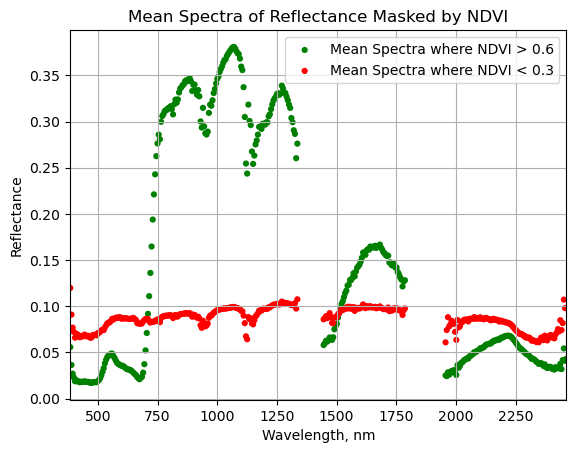
Plot the masked NDVI dataframe to display the mean spectra for NDVI values that exceed 0.6 and that are less than 0.3:
ax = plt.gca();
serc_ndvi_df.plot(ax=ax,x='wavelength',y='mean_refl_ndvi_gtpt6',color='green',
edgecolor='none',kind='scatter',label='Mean Spectra where NDVI > 0.6',legend=True);
serc_ndvi_df.plot(ax=ax,x='wavelength',y='mean_refl_ndvi_ltpt3',color='red',
edgecolor='none',kind='scatter',label='Mean Spectra where NDVI < 0.3',legend=True);
ax.set_title('Mean Spectra of Reflectance Masked by NDVI')
ax.set_xlim([np.nanmin(w),np.nanmax(w)]);
ax.set_xlabel("Wavelength, nm"); ax.set_ylabel("Reflectance")
ax.grid('on');