Tutorial
Introduction to NEON soil sensor data
Authors: Edward Ayres
Last Updated: Apr 10, 2025
This data tutorial provides instruction on working with three different NEON data products to investigate controls on soil CO2 concentrations:
- DP1.00041.001, Soil temperature
- DP1.00094.001, Soil water content and water salinity
- DP1.00095.001, Soil CO2 concentration
Soil temperature, soil water content, and soil CO2 concentration are measured in each of the five sensor-based soil plots at each NEON terrestrial site. Vertical profiles of soil temperature (up to 9 measurement levels per plot) and soil water content (up to 8 levels) are measured from near the soil surface down to 2 m deep or restrictive feature if shallower. Soil CO2 concentrations are measured at three different surface soil depths, typically <20 cm deep. Within each soil plot all these measurements are made within a few meters of one-another.
We will be using data from the Santa Rita Experimental Range (SRER) site in Arizona. The site is in the Sonoran Desert. Winters are short and mild, while summers are long and hot.
Things You’ll Need To Complete This Tutorial
You will need the most current version of R loaded on your computer to complete this tutorial.
1. Setup
Start by installing (if necessary) and loading the neonUtilities package.
Installation can be run once, then periodically to get package updates.
install.packages("neonUtilities")
Now load packages. This needs to be done every time you run code.
library(neonUtilities)
2. Download the data
Download the soil temperature, soil water content, and soil CO2 concentration
data using the loadByProduct() function in the neonUtilities package. Inputs
needed for the function are:
-
dpID: data product ID; soil temperature = DP1.00041.001 (or soil water content = DP1.00094.001, or soil CO2 concentration = DP1.00095.001) -
site: (vector of) 4-letter site codes; Santa Rita Experimental Range = SRER -
startdate: start year and month (YYYY-MM); January 2021 = 2021-01 -
enddate: end year and month (YYYY-MM); December 2021 = 2021-12 -
package: basic or expanded; we'll download basic here -
timeIndex: 1- or 30-minute averaging interval; we'll download 30-minute data -
check.size: should this function prompt the user with an estimated download size? Set toFALSEhere for ease of processing as a script, but good to leave as defaultTRUEwhen downloading a dataset for the first time.
Refer to the cheat sheet
for the neonUtilities package for more details if desired.
Note that this will download files totaling approximately 200 MB. If this is too large for your computer or internet connection you can reduce the date range and continue with the rest of the tutorial (e.g., startdate = 2021-06 and enddate = 2021-08; approximately 50 MB).
st <- loadByProduct(dpID="DP1.00041.001",
startdate="2021-01",
enddate="2021-12",
site="SRER",
package="basic",
timeIndex="30",
check.size=F)
swc <- loadByProduct(dpID="DP1.00094.001",
startdate="2021-01",
enddate="2021-12",
site="SRER",
package="basic",
timeIndex="30",
check.size=F)
co2 <- loadByProduct(dpID="DP1.00095.001",
startdate="2021-01",
enddate="2021-12",
site="SRER",
package="basic",
timeIndex="30",
check.size=F)
3. Soil temperature
The data we downloaded contains data from each of the five sensor-based soil plots, but here we will just focus on one of the soil plots (soil plot 1; horizontalPosition = "001"). In addition, soil temperature is measured at multiple depths in each soil plot, but for simplicity we will just use measurements with a nominal depth of 6 cm (verticalPosition = "502"). Lastly, we only want to use data that passed the QA/QC tests (finalQF = 0). The following steps will identify the rows that correspond to each of these conditions and we will then identify the rows corresponding to all of these conditions using the intersect() function.
p1rowsT <- grep("001", st$ST_30_minute$horizontalPosition)
d2rowsT <- grep("502", st$ST_30_minute$verticalPosition)
goodRowsT <- which(st$ST_30_minute$finalQF == 0)
useTheseT <- intersect(intersect(p1rowsT, d2rowsT), goodRowsT)
Next let's identify the exact measurement depth so we can add that to the plot legend. To do this we'll use the st$sensor_positions_00041 data frame, which contains information about the physical location of the sensors such as their depth, their distance from the soil plot reference corner, and the latitude, longitude and elevation of the soil plot reference corner. We can get a sense of the type of data in the sensor positions file using the head function.
head(st$sensor_positions_00041)
## siteID HOR.VER sensorLocationID sensorLocationDescription positionStartDateTime
## <char> <char> <char> <char> <char>
## 1: SRER 001.501 CFGLOC104513 Santa Rita Soil Temp Profile SP1, Z1 Depth 2010-01-01T00:00:00Z
## 2: SRER 001.502 CFGLOC104515 Santa Rita Soil Temp Profile SP1, Z2 Depth 2010-01-01T00:00:00Z
## 3: SRER 001.503 CFGLOC104518 Santa Rita Soil Temp Profile SP1, Z3 Depth 2010-01-01T00:00:00Z
## 4: SRER 001.504 CFGLOC104520 Santa Rita Soil Temp Profile SP1, Z4 Depth 2010-01-01T00:00:00Z
## 5: SRER 001.505 CFGLOC104522 Santa Rita Soil Temp Profile SP1, Z5 Depth 2010-01-01T00:00:00Z
## 6: SRER 001.506 CFGLOC104524 Santa Rita Soil Temp Profile SP1, Z6 Depth 2010-01-01T00:00:00Z
## positionEndDateTime referenceLocationID referenceLocationIDDescription
## <lgcl> <char> <char>
## 1: NA SOILPL104501 Santa Rita Soil Plot, SP1
## 2: NA SOILPL104501 Santa Rita Soil Plot, SP1
## 3: NA SOILPL104501 Santa Rita Soil Plot, SP1
## 4: NA SOILPL104501 Santa Rita Soil Plot, SP1
## 5: NA SOILPL104501 Santa Rita Soil Plot, SP1
## 6: NA SOILPL104501 Santa Rita Soil Plot, SP1
## referenceLocationIDStartDateTime referenceLocationIDEndDateTime xOffset yOffset zOffset pitch roll
## <char> <lgcl> <num> <num> <num> <num> <int>
## 1: 2010-01-01T00:00:00Z NA 0.97 2.7 -0.02 0.6 0
## 2: 2010-01-01T00:00:00Z NA 0.97 2.7 -0.06 0.6 0
## 3: 2010-01-01T00:00:00Z NA 0.97 2.7 -0.16 0.6 0
## 4: 2010-01-01T00:00:00Z NA 0.97 2.7 -0.26 0.6 0
## 5: 2010-01-01T00:00:00Z NA 0.97 2.7 -0.56 0.6 0
## 6: 2010-01-01T00:00:00Z NA 0.97 2.7 -0.96 0.6 0
## azimuth locationReferenceLatitude locationReferenceLongitude locationReferenceElevation eastOffset
## <int> <num> <num> <num> <num>
## 1: 30 31.91062 -110.8353 999.36 -1.85
## 2: 30 31.91062 -110.8353 999.36 -1.85
## 3: 30 31.91062 -110.8353 999.36 -1.85
## 4: 30 31.91062 -110.8353 999.36 -1.85
## 5: 30 31.91062 -110.8353 999.36 -1.85
## 6: 30 31.91062 -110.8353 999.36 -1.85
## northOffset xAzimuth yAzimuth publicationDate
## <num> <int> <int> <char>
## 1: 2.19 30 300 20221210T203358Z
## 2: 2.19 30 300 20221210T203358Z
## 3: 2.19 30 300 20221210T203358Z
## 4: 2.19 30 300 20221210T203358Z
## 5: 2.19 30 300 20221210T203358Z
## 6: 2.19 30 300 20221210T203358Z
We just want to know the depth (zOffset) of the sensor at soil plot 1 measurement level 2 (HOR.VER = "001.502") so we'll filter that value.
st$sensor_positions_00041[grep("001.502", st$sensor_positions_00041$HOR.VER), "zOffset"]
## zOffset
## <num>
## 1: -0.06
This shows a zOffset of -0.06, indicating that the measurement was 0.06 m (6 cm) below the soil surface. Now let's see what the data look like! Make a time series plot of soil temperature at SRER soil plot 1 measurement level 2 and add a legend indicating the sensor was at 6 cm.
plot(st$ST_30_minute$startDateTime[useTheseT],
st$ST_30_minute$soilTempMean[useTheseT],
pch=".",
xlab="",
ylab="Soil temperature (°C)",
main="SRER soil plot 1, 2021")
legend("topleft", legend="6 cm", lty=1, bty="n")
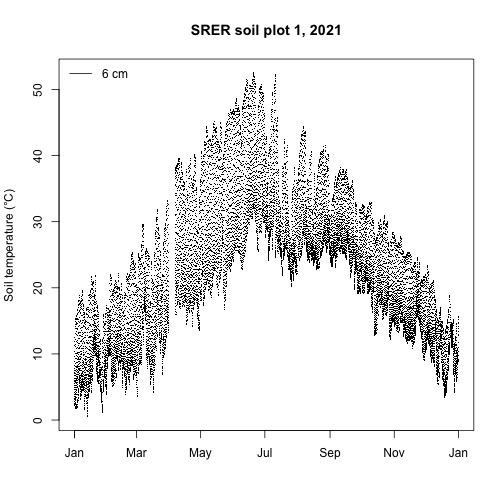
We see the expected pattern of warmer soil temperatures in the summer and cooler temperatures in the winter along with the typical diurnal cycles.
4. Soil water content
Now we'll identify the soil water content rows corresponding to soil plot 1 (horizontalPosition = "001"), with a nominal depth of 6 cm (verticalPosition = "501"), and that passed the QA/QC tests (VSWCFinalQF = 0).
p1rowsM <- grep("001", swc$SWS_30_minute$horizontalPosition)
d1rowsM <- grep("501", swc$SWS_30_minute$verticalPosition)
goodRowsM <- which(swc$SWS_30_minute$VSWCFinalQF == 0)
useTheseM <- intersect(intersect(p1rowsM, d1rowsM), goodRowsM)
Let's create a time series plot of soil water content based on this data, but first we'll use the expression() function to create an axis label that contains superscripts for the units.
The soil water content data product page tells us that there is currently a problem with the sensor depths in the sensor_positions table in the soil water content data product and that we should instead use a file called swc_depthsV2.csv to identify the correct depths, which can be downloaded from the Documentation section of the webpage. Depths are not currently displayed correctly in the sensor_positions table because the raw sensor data includes all measurement levels in a single data stream and the data processing pipeline is not capable of storing multiple measurement depths for a single data stream. Future upgrades to the data processing pipeline will resolve this problem.
Looking at the row in this file with siteID = SRER, horizontalPosition.HOR = "001" (soil plot 1), and verticalPosition.VER = "501 (measurement level 1) we see that the measurement depth was -0.06 m (i.e., 6 cm below the soil surface). Now we can also add a legend to the soil water content time series.
labelM=expression(paste("Soil water content (m"^" 3", " m"^"-3", ")"))
plot(swc$SWS_30_minute$startDateTime[useTheseM],
swc$SWS_30_minute$VSWCMean[useTheseM],
pch=".",
xlab="",
ylab=labelM)
legend("topleft", legend="6 cm", lty=1, bty="n")
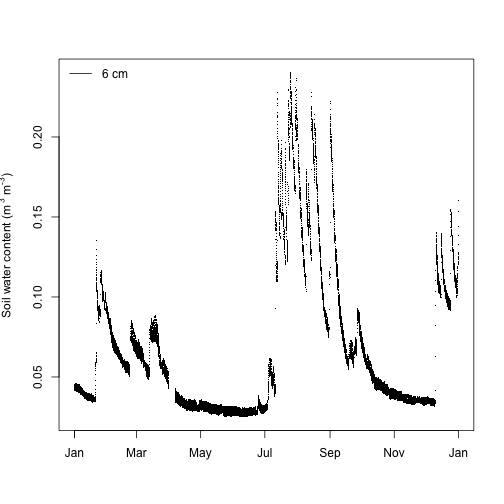
Looks good except the superscripts are partially cut off on the y-axis label. Let's adjust the plot margins to create enough space for the label using the mar graphical parameter.
par(mar=c(3,5,2,1))
plot(swc$SWS_30_minute$startDateTime[useTheseM],
swc$SWS_30_minute$VSWCMean[useTheseM],
pch=".",
xlab="",
ylab=labelM)
legend("topleft", legend="6 cm", lty=1, bty="n")
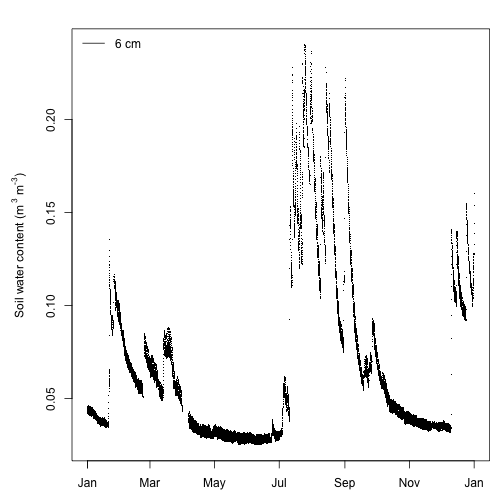
That's better. Soil water content shows typical patterns of sharp rises in moisture (presumably from rain events) followed by gradual declines as the soil dries. Soil moisture has a bimodal distribution being higher during winter and late summer, which is consistent with meteorology at SRER with winter rain as well as late summer thunderstorms.
5. Soil CO2 concentration
Now we want to see the temporal patterns in soil CO2 concentration data. Soil CO2 concentration is measured at three depths in surface soils in each soil plot and we'll look at data from all three depths. As we did with soil temperature and soil water content, we'll first identify the rows that correspond to soil plot 1, each measurement level (verticalPosition = "501", "502", or "503"), and that passed the QA/QC tests (finalQF = 0).
# Identify rows for soil plot 1
p1rowsC <- grep("001", co2$SCO2C_30_minute$horizontalPosition)
# Identify rows for measurement levels 1, 2, and 3
d1rowsC <- grep("501", co2$SCO2C_30_minute$verticalPosition)
d2rowsC <- grep("502", co2$SCO2C_30_minute$verticalPosition)
d3rowsC <- grep("503", co2$SCO2C_30_minute$verticalPosition)
# Identify rows that passed the QA/QC tests
goodRowsC <- which(co2$SCO2C_30_minute$finalQF == 0)
# Identify rows for soil plot 1 that passed the QA/QC tests for each measurement level
useTheseC1 <- intersect(intersect(p1rowsC, d1rowsC), goodRowsC)
useTheseC2 <- intersect(intersect(p1rowsC, d2rowsC), goodRowsC)
useTheseC3 <- intersect(intersect(p1rowsC, d3rowsC), goodRowsC)
Next let's find out the depths of these measurements by identifying the rows corresponding to soil plot 1 in the sensor_positions table.
rows <- grep(c("001"), co2$sensor_positions_00095$HOR.VER)
co2$sensor_positions_00095[rows, c("zOffset")]
## zOffset
## <num>
## 1: -0.02
## 2: -0.05
## 3: -0.19
The sensors were measuring at 2, 5, and 19 cm below the soil surface.
Next we'll create an axis label using the expression() function to display the subscript and then create the time series plot. The points() function is used to add data from measurement levels 2 and 3 to the graph of level 1.
labelC=expression(paste("Soil CO"[2]," concentration (ppm)"))
plot(co2$SCO2C_30_minute$startDateTime[useTheseC1],
co2$SCO2C_30_minute$soilCO2concentrationMean[useTheseC1],
pch=".",
xlab="",
ylab=labelC,
ylim=c(0, 10000))
points(co2$SCO2C_30_minute$startDateTime[useTheseC2],
co2$SCO2C_30_minute$soilCO2concentrationMean[useTheseC2],
pch=".",
col="red")
points(co2$SCO2C_30_minute$startDateTime[useTheseC3],
co2$SCO2C_30_minute$soilCO2concentrationMean[useTheseC3],
pch=".",
col="blue")
legend("topleft", legend=c("2 cm", "5 cm", "19 cm"), lty=1, col=c("black", "red", "blue"), bty="n")
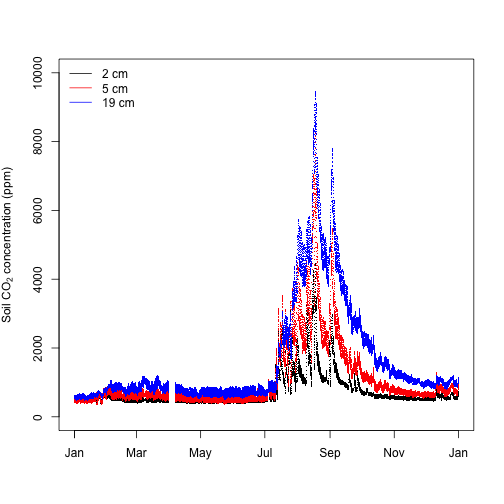
Soil CO2 concentrations were higher in the late summer and early fall and close to atmospheric levels in the winter through to early summer, likely reflecting periods of higher root, microbial and other soil organism activity. The typical soil CO2 concentration depth profile is also clear, with higher concentrations deeper in the soil reflecting the long time it takes CO2 produced at depth to diffuse to the atmosphere relative to CO2 produced near the soil surface.
6. Displaying the time series together
Now we've created separate time series plots for soil temperature, water content, and soil CO2 concentration. However, to help us looks for relationships between the three data sets it can be useful to plot them all on a single multi-panel plot.
To do this we will first change the graphical parameter mfcol to produce one column with three rows (one row for each plot). We
will also change the margins of the plots by adjusting the mar parameter to leave enough space for the axis labels. Then we'll add the temperature, water content, and CO2 concentration plots sequentially using the same code as above.
par(mfcol=c(3,1))
par(mar=c(3,5,2,1))
# Add soil temperature plot
plot(st$ST_30_minute$startDateTime[useTheseT],
st$ST_30_minute$soilTempMean[useTheseT],
pch=".",
xlab="",
ylab="Soil temperature (°C)",
main="SRER soil plot 1, 2021")
legend("topleft", legend="6 cm", lty=1, bty="n")
# Add soil water content plot
plot(swc$SWS_30_minute$startDateTime[useTheseM],
swc$SWS_30_minute$VSWCMean[useTheseM],
pch=".",
xlab="",
ylab=labelM)
legend("topleft", legend="6 cm", lty=1, bty="n")
# Add soil CO2 concentration plot
plot(co2$SCO2C_30_minute$startDateTime[useTheseC1],
co2$SCO2C_30_minute$soilCO2concentrationMean[useTheseC1],
pch=".",
xlab="",
ylab=labelC,
ylim=c(0, 10000))
points(co2$SCO2C_30_minute$startDateTime[useTheseC2],
co2$SCO2C_30_minute$soilCO2concentrationMean[useTheseC2],
pch=".",
col="red")
points(co2$SCO2C_30_minute$startDateTime[useTheseC3],
co2$SCO2C_30_minute$soilCO2concentrationMean[useTheseC3],
pch=".",
col="blue")
legend("topleft", legend=c("2 cm", "5 cm", "19 cm"), lty=1, col=c("black", "red", "blue"), bty="n")
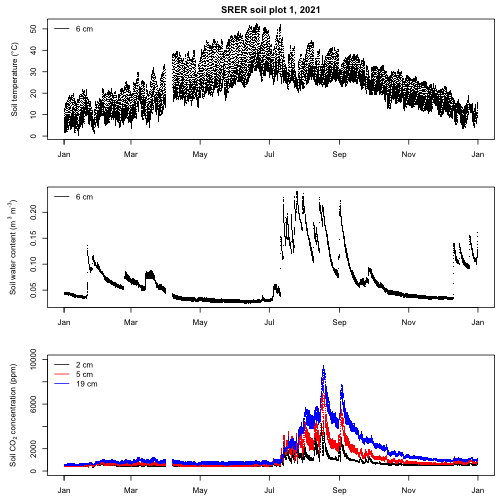
This multi-panel plot suggests that both soil temperature and water content influence soil CO2 concentrations at SRER. Specifically, soil CO2 concentrations tend to be low when the soil is cool regardless of water content, likewise concentrations tend to be low when the soil is dry regardless of temperature. When the soil is warm, CO2 concentrations responds rapidly to increases in soil moisture and then gradually decrease as the soil dries, presumably due to changes in the activity of roots and soil organisms.
In this tutorial we've focused on soil CO2 concentrations but most researchers are more interested in soil respiration rates than the soil CO2 concentrations themselves. Soil respiration can be calculated using these data products in combination with other NEON products, but this requires calculation of the soil CO2 diffusivity coefficient which is too complex to include in a brief data skills tutorial. However, some researchers have already started developing code to make these calculations based on NEON data (e.g., Ayres 2019, Zobitz et al. 2022a, Zobitz et al. 2022b).