Workshop
Access & Use of NEON Data, Aquatics Training Palooza 2018
NEON
Through participation in this workshop, full-time field ecologists on the NEON project will improve their ability to work with the local scientific community (site hosts, local researchers, and other potential NEON data users) by increasing their knowledge of the online resources available from NEON and the skills needed to access NEON data. In addition, the workshop will enable the field ecologists to directly work with NEON data by teaching them basic data skills that will increase their ability to conduct basic QA/QC on the data.
- Explain to site hosts and researchers at the sites where to go for more information on NEON, NEON data collection methods, NEON data, and training on using NEON data (neonscience.org).
- Explain to site hosts and researchers at field sites where to find information on what data is available for download (data.neonscience.org).
- Explain to potential data users multiple methods for accessing NEON data:
- through NEON data portal
- direct API calls
- through software packages, like the R package nneo
- Download data for a specified data product, site, and time frame from the data portal.
- Use the neonstackR package to combine multiple site/months .csv files to get a complete data set of interest.
- Produce line/bar plots of data products of interest.
Things You’ll Need To Complete This Workshop
1) Install/Update Software
To participant in this workshop, you will need a laptop with the most current version of R, and, preferably, RStudio loaded on your computer. For details on installing RStudio in Mac, PC, or Linux operating systems, please see the Additional Set Up Instructions section at the bottom of this page.
Please note, while these packages are required, a working knowledge of using R and RStudio is not required.
If you already have these installed on your laptop, please be sure that you are running the most current version of RStudio, R and all packages that we'll be using in the workshop (listed below).
2) Install R Packages
We will be using the package neonDataStackR in this workshop. To install, open up RStudio, in the bottom left window of RStudio -- the "console" type in the following set of commands.
install.packages("devtools")
library(devtools)
devtools::install_github("NEONScience/NEON-utilities/neonDataStackR")
Then to make sure the installation worked:
library(neonDataStackR)
If you get no output, it is correctly installed. If you get an error, please repeat the steps.
If you already have neonDataStackR installed, please update you version of neonDataStackR as it is frequently updated.
3) Data to Download
Assuming there are no internet problems, we will be using data directly from the portal. However, please download this sample data file so that, if internet issues arise during the workshop, we will have data to work with.
Download NEON Surface Water Chemistry Data SubsetPlease keep the downloaded data in your "Downloads" folder. This data is downloaded as back up in case there are issues with the internet connectivity or data portal during the workshop.
Workshop Instructor
- Megan A. Jones; @meganahjones; Staff Scientists/Science Educator, NEON
Please get in touch with the instructor prior to the workshop with any questions.
Twitter?
Please tweet @NEON_sci and with the #WorkWithData hashtag during the workshop.
Schedule
Please note that the schedule listed below may change depending upon the pace of the workshop!
| Time | Topic |
|---|---|
| 14:00 | Information for Understanding NEON: neonscience.org |
| 14:30 | Accessing NEON data: data.neonscience.org |
| Download via the Data Portal | |
| Access through the NEON API | |
| Access using software packages, like nneo R package | |
| 15:30 | Using the neonDataStackR script in R |
| 16:00 | --------- BREAK --------- |
| 16:15 | Working with NEON data products |
| 17:45 | Data Challenge |
Additional Set Up Resources
[[nid:6408]]
| Time | Topic |
|---|---|
| 14:00 | Information for Understanding NEON: neonscience.org |
| 14:30 | Accessing NEON data: data.neonscience.org |
| Download via the Data Portal | |
| Access through the NEON API | |
| Access using software packages, like nneo R package | |
| 15:30 | Using the neonDataStackR script in R |
| 16:00 | --------- BREAK --------- |
| 16:15 | Working with NEON data products |
| 17:45 | Data Challenge |
Related Tutorials
Use the neonUtilities Package to Access NEON Data
This tutorial provides an overview of functions in the
neonUtilities package in R and the
neonutilities package in Python. These packages provide a
toolbox of basic functionality for working with NEON data.
This tutorial is primarily an index of functions and their inputs;
for more in-depth guidance in using these functions to work with NEON
data, see the
Download
and Explore tutorial. If you are already familiar with the
neonUtilities package, and need a quick reference guide to
function inputs and notation, see the
neonUtilities
cheat sheet.
Function index
The neonUtilities/neonutilities package
contains several functions (use the R and Python tabs to see the syntax
in each language):
R
stackByTable(): Takes zip files downloaded from the Data Portal or downloaded byzipsByProduct(), unzips them, and joins the monthly files by data table to create a single file per table.zipsByProduct(): A wrapper for the NEON API; downloads data based on data product and site criteria. Stores downloaded data in a format that can then be joined bystackByTable().loadByProduct(): Combines the functionality ofzipsByProduct(),
stackByTable(), andreadTableNEON(): Downloads the specified data, stacks the files, and loads the files to the R environment.byFileAOP(): A wrapper for the NEON API; downloads remote sensing data based on data product, site, and year criteria. Preserves the file structure of the original data.byTileAOP(): Downloads remote sensing data for the specified data product, subset to tiles that intersect a list of coordinates.readTableNEON(): Reads NEON data tables into R, using the variables file to assign R classes to each column.getCitation(): Get a BibTeX citation for a particular data product and release.
Python
stack_by_table(): Takes zip files downloaded from the Data Portal or downloaded byzips_by_product(), unzips them, and joins the monthly files by data table to create a single file per table.zips_by_product(): A wrapper for the NEON API; downloads data based on data product and site criteria. Stores downloaded data in a format that can then be joined bystack_by_table().load_by_product(): Combines the functionality ofzips_by_product(),
stack_by_table(), andread_table_neon(): Downloads the specified data, stacks the files, and loads the files to the R environment.by_file_aop(): A wrapper for the NEON API; downloads remote sensing data based on data product, site, and year criteria. Preserves the file structure of the original data.by_tile_aop(): Downloads remote sensing data for the specified data product, subset to tiles that intersect a list of coordinates.read_table_neon(): Reads NEON data tables into R, using the variables file to assign R classes to each column.get_citation(): Get a BibTeX citation for a particular data product and release.
If you are only interested in joining data
files downloaded from the NEON Data Portal, you will only need to use
stackByTable(). Follow the instructions in the first
section of the
Download
and Explore tutorial.
Install and load packages
First, install and load the package. The installation step only needs to be run once, and then periodically to update when new package versions are released. The load step needs to be run every time you run your code.
R
##
## # install neonUtilities - can skip if already installed
## install.packages("neonUtilities")
##
## # load neonUtilities
library(neonUtilities)
## Python
# install neonutilities - can skip if already installed
# do this in the command line
pip install neonutilities
# load neonutilities in working environment
import neonutilities as nuDownload files and load to working environment
The most popular function in neonUtilities is
loadByProduct() (or load_by_product() in
neonutilities). This function downloads data from the NEON
API, merges the site-by-month files, and loads the resulting data tables
into the programming environment, classifying each variable’s data type
appropriately. It combines the actions of the
zipsByProduct(), stackByTable(), and
readTableNEON() functions, described below.
This is a popular choice because it ensures you’re always working with the latest data, and it ends with ready-to-use tables. However, if you use it in a workflow you run repeatedly, keep in mind it will re-download the data every time.
loadByProduct() works on most observational (OS) and
sensor (IS) data, but not on surface-atmosphere exchange (SAE) data,
remote sensing (AOP) data, and some of the data tables in the microbial
data products. For functions that download AOP data, see the
byFileAOP() and byTileAOP() sections in this
tutorial. For functions that work with SAE data, see the
NEON
eddy flux data tutorial. SAE functions are not yet available in
Python.
The inputs to loadByProduct() control which data to
download and how to manage the processing:
R
dpID: The data product ID, e.g. DP1.00002.001site: Defaults to “all”, meaning all sites with available data; can be a vector of 4-letter NEON site codes, e.g.c("HARV","CPER","ABBY").startdateandenddate: Defaults to NA, meaning all dates with available data; or a date in the form YYYY-MM, e.g. 2017-06. Since NEON data are provided in month packages, finer scale querying is not available. Both start and end date are inclusive.package: Either basic or expanded data package. Expanded data packages generally include additional information about data quality, such as chemical standards and quality flags. Not every data product has an expanded package; if the expanded package is requested but there isn’t one, the basic package will be downloaded.timeIndex: Defaults to “all”, to download all data; or the number of minutes in the averaging interval. See example below; only applicable to IS data.release: Specify a particular data Release, e.g."RELEASE-2024". Defaults to the most recent Release. For more details and guidance, see the Release and Provisional tutorial.include.provisional: T or F: Should provisional data be downloaded? Ifreleaseis not specified, set to T to include provisional data in the download. Defaults to F.savepath: the file path you want to download to; defaults to the working directory.check.size: T or F: should the function pause before downloading data and warn you about the size of your download? Defaults to T; if you are using this function within a script or batch process you will want to set it to F.token: Optional API token for faster downloads. See the API token tutorial.nCores: Number of cores to use for parallel processing. Defaults to 1, i.e. no parallelization.
Python
dpid: the data product ID, e.g. DP1.00002.001site: defaults to “all”, meaning all sites with available data; can be a list of 4-letter NEON site codes, e.g.["HARV","CPER","ABBY"].startdateandenddate: defaults to NA, meaning all dates with available data; or a date in the form YYYY-MM, e.g. 2017-06. Since NEON data are provided in month packages, finer scale querying is not available. Both start and end date are inclusive.package: either basic or expanded data package. Expanded data packages generally include additional information about data quality, such as chemical standards and quality flags. Not every data product has an expanded package; if the expanded package is requested but there isn’t one, the basic package will be downloaded.timeindex: defaults to “all”, to download all data; or the number of minutes in the averaging interval. See example below; only applicable to IS data.release: Specify a particular data Release, e.g."RELEASE-2024". Defaults to the most recent Release. For more details and guidance, see the Release and Provisional tutorial.include_provisional: True or False: Should provisional data be downloaded? Ifreleaseis not specified, set to T to include provisional data in the download. Defaults to F.savepath: the file path you want to download to; defaults to the working directory.check_size: True or False: should the function pause before downloading data and warn you about the size of your download? Defaults to True; if you are using this function within a script or batch process you will want to set it to False.token: Optional API token for faster downloads. See the API token tutorial.cloud_mode: Can be set to True if you are working in a cloud environment; provides more efficient data transfer from NEON cloud storage to other cloud environments.progress: Set to False to omit the progress bar during download and stacking.
The dpID (dpid) is the data product
identifier of the data you want to download. The DPID can be found on
the
Explore Data Products page. It will be in the form DP#.#####.###
Demo data download and read
Let’s get triple-aspirated air temperature data (DP1.00003.001) from
Moab and Onaqui (MOAB and ONAQ), from May–August 2018, and name the data
object triptemp:
R
triptemp <- loadByProduct(dpID="DP1.00003.001",
site=c("MOAB","ONAQ"),
startdate="2018-05",
enddate="2018-08")Python
triptemp = nu.load_by_product(dpid="DP1.00003.001",
site=["MOAB","ONAQ"],
startdate="2018-05",
enddate="2018-08")View downloaded data
The object returned by loadByProduct() is a named list
of data tables, or a dictionary of data tables in Python. To work with
each of them, select them from the list.
R
names(triptemp)## [1] "citation_00003_RELEASE-2024" "issueLog_00003"
## [3] "readme_00003" "sensor_positions_00003"
## [5] "TAAT_1min" "TAAT_30min"
## [7] "variables_00003"temp30 <- triptemp$TAAT_30minIf you prefer to extract each table from the list and work with it as
an independent object, you can use the list2env()
function:
list2env(trip.temp, .GlobalEnv)Python
triptemp.keys()## dict_keys(['TAAT_1min', 'TAAT_30min', 'citation_00003_RELEASE-2024', 'issueLog_00003', 'readme_00003', 'sensor_positions_00003', 'variables_00003'])temp30 = triptemp["TAAT_30min"]If you prefer to extract each table from the list and work with it as
an independent object, you can use
globals().update():
globals().update(triptemp)For more details about the contents of the data tables and metadata tables, check out the Download and Explore tutorial.
Join data files: stackByTable()
The function stackByTable() joins the month-by-site
files from a data download. The output will yield data grouped into new
files by table name. For example, the single aspirated air temperature
data product contains 1 minute and 30 minute interval data. The output
from this function is one .csv with 1 minute data and one .csv with 30
minute data.
Depending on your file size this function may run for a while. For example, in testing for this tutorial, 124 MB of temperature data took about 4 minutes to stack. A progress bar will display while the stacking is in progress.
Download the Data
To stack data from the Portal, first download the data of interest
from the NEON
Data Portal. To stack data downloaded from the API, see the
zipsByProduct() section below.
Your data will download from the Portal in a single zipped file.
The stacking function will only work on zipped Comma Separated Value (.csv) files and not the NEON data stored in other formats (HDF5, etc).
Run stackByTable()
The example data below are single-aspirated air temperature.
To run the stackByTable() function, input the file path
to the downloaded and zipped file.
R
# Modify the file path to the file location on your computer
stackByTable(filepath="~neon/data/NEON_temp-air-single.zip")Python
# Modify the file path to the file location on your computer
nu.stack_by_table(filepath="/neon/data/NEON_temp-air-single.zip")In the same directory as the zipped file, you should now have an unzipped directory of the same name. When you open this you will see a new directory called stackedFiles. This directory contains one or more .csv files (depends on the data product you are working with) with all the data from the months & sites you downloaded. There will also be a single copy of the associated variables, validation, and sensor_positions files, if applicable (validation files are only available for observational data products, and sensor position files are only available for instrument data products).
These .csv files are now ready for use with the program of your choice.
To read the data tables, we recommend using
readTableNEON(), which will assign each column to the
appropriate data type, based on the metadata in the variables file. This
ensures time stamps and missing data are interpreted correctly.
Load data to environment
R
SAAT30 <- readTableNEON(
dataFile='~/stackedFiles/SAAT_30min.csv',
varFile='~/stackedFiles/variables_00002.csv'
)Python
SAAT30 = nu.read_table_neon(
dataFile='/stackedFiles/SAAT_30min.csv',
varFile='/stackedFiles/variables_00002.csv'
)Other function inputs
Other input options in stackByTable() are:
savepath: allows you to specify the file path where you want the stacked files to go, overriding the default. Set to"envt"to load the files to the working environment.saveUnzippedFiles: allows you to keep the unzipped, unstacked files from an intermediate stage of the process; by default they are discarded.
Example usage:
R
stackByTable(filepath="~neon/data/NEON_temp-air-single.zip",
savepath="~data/allTemperature", saveUnzippedFiles=T)
tempsing <- stackByTable(filepath="~neon/data/NEON_temp-air-single.zip",
savepath="envt", saveUnzippedFiles=F)
Python
nu.stack_by_table(filepath="/neon/data/NEON_temp-air-single.zip",
savepath="/data/allTemperature",
saveUnzippedFiles=True)
tempsing <- nu.stack_by_table(filepath="/neon/data/NEON_temp-air-single.zip",
savepath="envt",
saveUnzippedFiles=False)
Download files to be stacked: zipsByProduct()
The function zipsByProduct() is a wrapper for the NEON
API, it downloads zip files for the data product specified and stores
them in a format that can then be passed on to
stackByTable().
Input options for zipsByProduct() are the same as those
for loadByProduct() described above.
Here, we’ll download single-aspirated air temperature (DP1.00002.001) data from Wind River Experimental Forest (WREF) for April and May of 2019.
R
zipsByProduct(dpID="DP1.00002.001", site="WREF",
startdate="2019-04", enddate="2019-05",
package="basic", check.size=T)Downloaded files can now be passed to stackByTable() to
be stacked.
stackByTable(filepath=paste(getwd(),
"/filesToStack00002",
sep=""))Python
nu.zips_by_product(dpid="DP1.00002.001", site="WREF",
startdate="2019-04", enddate="2019-05",
package="basic", check_size=True)Downloaded files can now be passed to stackByTable() to
be stacked.
nu.stack_by_table(filepath=os.getcwd()+
"/filesToStack00002")For many sensor data products, download sizes can get very large, and
stackByTable() takes a long time. The 1-minute or 2-minute
files are much larger than the longer averaging intervals, so if you
don’t need high- frequency data, the timeIndex input option
lets you choose which averaging interval to download.
This option is only applicable to sensor (IS) data, since OS data are not averaged.
Download by averaging interval
Download only the 30-minute data for single-aspirated air temperature at WREF:
R
zipsByProduct(dpID="DP1.00002.001", site="WREF",
startdate="2019-04", enddate="2019-05",
package="basic", timeIndex=30,
check.size=T)Python
nu.zips_by_product(dpid="DP1.00002.001", site="WREF",
startdate="2019-04",
enddate="2019-05", package="basic",
timeindex=30, check_size=True)The 30-minute files can be stacked and loaded as usual.
Download remote sensing files
Remote sensing data files can be very large, and NEON remote sensing
(AOP) data are stored in a directory structure that makes them easier to
navigate. byFileAOP() downloads AOP files from the API
while preserving their directory structure. This provides a convenient
way to access AOP data programmatically.
Be aware that downloads from byFileAOP() can take a VERY
long time, depending on the data you request and your connection speed.
You may need to run the function and then leave your machine on and
downloading for an extended period of time.
Here the example download is the Ecosystem Structure data product at Hop Brook (HOPB) in 2017; we use this as the example because it’s a relatively small year-site-product combination.
R
byFileAOP("DP3.30015.001", site="HOPB",
year=2017, check.size=T)Python
nu.by_file_aop(dpid="DP3.30015.001",
site="HOPB", year=2017,
check_size=True)The files should now be downloaded to a new folder in your working directory.
Download remote sensing files for specific coordinates
Often when using remote sensing data, we only want data covering a
certain area - usually the area where we have coordinated ground
sampling. byTileAOP() queries for data tiles containing a
specified list of coordinates. It only works for the tiled, AKA
mosaicked, versions of the remote sensing data, i.e. the ones with data
product IDs beginning with “DP3”.
Here, we’ll download tiles of vegetation indices data (DP3.30026.001) corresponding to select observational sampling plots. For more information about accessing NEON spatial data, see the API tutorial and the in-development geoNEON package.
For now, assume we’ve used the API to look up the plot centroids of plots SOAP_009 and SOAP_011 at the Soaproot Saddle site. You can also look these up in the Spatial Data folder of the document library. The coordinates of the two plots in UTMs are 298755,4101405 and 299296,4101461. These are 40x40m plots, so in looking for tiles that contain the plots, we want to include a 20m buffer. The “buffer” is actually a square, it’s a delta applied equally to both the easting and northing coordinates.
R
byTileAOP(dpID="DP3.30026.001", site="SOAP",
year=2018, easting=c(298755,299296),
northing=c(4101405,4101461),
buffer=20)Python
nu.by_tile_aop(dpid="DP3.30026.001",
site="SOAP", year=2018,
easting=[298755,299296],
northing=[4101405,4101461],
buffer=20)The 2 tiles covering the SOAP_009 and SOAP_011 plots have
been downloaded.
Using the NEON API in R
This is a tutorial in pulling data from the NEON API or Application Programming Interface. The tutorial uses R and the R package httr, but the core information about the API is applicable to other languages and approaches.
Objectives
After completing this activity, you will be able to:
- Construct API calls to query the NEON API.
- Access and understand data and metadata available via the NEON API.
Things You’ll Need To Complete This Tutorial
To complete this tutorial you will need the most current version of R and, preferably, RStudio loaded on your computer.
Install R Packages
-
httr:
install.packages("httr") -
jsonlite:
install.packages("jsonlite")
Additional Resources
What is an API?
If you are unfamiliar with the concept of an API, think of an API as a ‘middle person' that provides a communication path for a software application to obtain information from a digital data source. APIs are becoming a very common means of sharing digital information. Many of the apps that you use on your computer or mobile device to produce maps, charts, reports, and other useful forms of information pull data from multiple sources using APIs. In the ecological and environmental sciences, many researchers use APIs to programmatically pull data into their analyses. (Quoted from the NEON Observatory Blog story: API and data availability viewer now live on the NEON data portal.)
What is accessible via the NEON API?
The NEON API includes endpoints for NEON data and metadata, including spatial data, taxonomic data, and samples (see Endpoints below). This tutorial explores these sources of information using a specific data product as a guide. The principles and rule sets described below can be applied to other data products and metadata.
Anatomy of an API call
An example API call: http://data.neonscience.org/api/v0/data/DP1.10003.001/WOOD/2015-07
This includes the base URL, endpoint, and target.
Base URL:
http://data.neonscience.org/api/v0/data/DP1.10003.001/WOOD/2015-07
Specifics are appended to this in order to get the data or metadata you're looking for, but all calls to an API will include the base URL. For the NEON API, this is http://data.neonscience.org/api/v0 -- not clickable, because the base URL by itself will take you nowhere!
Endpoints:
http://data.neonscience.org/api/v0/data/DP1.10098.001/WOOD/2015-07
What type of data or metadata are you looking for?
-
~/products Information about one or all of NEON's data products
-
~/sites Information about data availability at the site specified in the call
-
~/locations Spatial data for the NEON locations specified in the call
-
~/data Data! By product, site, and date (in monthly chunks)
-
~/samples Information about sample relationships and sample tracking
-
~/taxonomy Access to NEON's taxon lists, the approved scientific names for sampled taxa
Targets:
http://data.neonscience.org/api/v0/data/DP1.10098.001/WOOD/2015-07
The specific data product, location, sample, etc, you want to get data for.
Data, by way of Products
Which product do you want to get data for? Consult the Explore Data Products page.
We'll pick Woody vegetation structure, DP1.10098.001
Your first thought is probably to use the /data endpoint. And we'll get there. But notice above that the API call for the /data endpoint includes the site and month of data to download. You don't want to have to guess sites and months at random - first, you need to see which sites and months have available data for the product you're interested in. That can be done either through the /sites or the /products endpoint; here we'll use /products.
Note: Checking for data availability can sometimes be skipped for the streaming sensor data products. In general, they are available continuously, and you could theoretically query a site and month of interest and expect there to be data by default. However, there can be interruptions to sensor data, in particular at aquatic sites, so checking availability first is the most reliable approach.
Use the products endpoint to query for Woody vegetation data. The target is the data product identifier, noted above, DP1.10098.001:
# Load the necessary libraries
library(httr)
library(jsonlite)
# Request data using the GET function & the API call
req <- GET("http://data.neonscience.org/api/v0/products/DP1.10098.001")
req
## Response [https://data.neonscience.org/api/v0/products/DP1.10098.001]
## Date: 2021-06-16 01:03
## Status: 200
## Content-Type: application/json;charset=UTF-8
## Size: 70.1 kB
The object returned from GET() has many layers of information. Entering the
name of the object gives you some basic information about what you accessed.
Status: 200 indicates this was a successful query; the status field can be
a useful place to look if something goes wrong. These are HTTP status codes,
you can google them to find out what a given value indicates.
The Content-Type parameter tells us we've accessed a json file. The easiest
way to translate this to something more manageable in R is to use the
fromJSON() function in the jsonlite package. It will convert the json into
a nested list, flattening the nesting where possible.
# Make the data readable by jsonlite
req.text <- content(req, as="text")
# Flatten json into a nested list
avail <- jsonlite::fromJSON(req.text,
simplifyDataFrame=T,
flatten=T)
A lot of the content here is basic information about the data product.
You can see all of it by running the line print(avail), but
this will result in a very long printout in your console. Instead, try viewing
list items individually. Here, we highlight a couple of interesting examples:
# View description of data product
avail$data$productDescription
## [1] "Structure measurements, including height, crown diameter, and stem diameter, as well as mapped position of individual woody plants"
# View data product abstract
avail$data$productAbstract
## [1] "This data product contains the quality-controlled, native sampling resolution data from in-situ measurements of live and standing dead woody individuals and shrub groups, from all terrestrial NEON sites with qualifying woody vegetation. The exact measurements collected per individual depend on growth form, and these measurements are focused on enabling biomass and productivity estimation, estimation of shrub volume and biomass, and calibration / validation of multiple NEON airborne remote-sensing data products. In general, comparatively large individuals that are visible to remote-sensing instruments are mapped, tagged and measured, and other smaller individuals are tagged and measured but not mapped. Smaller individuals may be subsampled according to a nested subplot approach in order to standardize the per plot sampling effort. Structure and mapping data are reported per individual per plot; sampling metadata, such as per growth form sampling area, are reported per plot. For additional details, see the user guide, protocols, and science design listed in the Documentation section in this data product's details webpage.\n\nLatency:\nThe expected time from data and/or sample collection in the field to data publication is as follows, for each of the data tables (in days) in the downloaded data package. See the Data Product User Guide for more information.\n\nvst_apparentindividual: 90\n\nvst_mappingandtagging: 90\n\nvst_perplotperyear: 300\n\nvst_shrubgroup: 90"
You may notice that some of this information is also accessible on the NEON data portal. The portal uses the same data sources as the API, and in many cases the portal is using the API on the back end, and simply adding a more user-friendly display to the data.
We want to find which sites and months have available data. That is in the
siteCodes section. Let's look at what information is presented for each
site:
# Look at the first list element for siteCode
avail$data$siteCodes$siteCode[[1]]
## [1] "ABBY"
# And at the first list element for availableMonths
avail$data$siteCodes$availableMonths[[1]]
## [1] "2015-07" "2015-08" "2016-08" "2016-09" "2016-10" "2016-11" "2017-03" "2017-04" "2017-07" "2017-08"
## [11] "2017-09" "2018-07" "2018-08" "2018-09" "2018-10" "2018-11" "2019-07" "2019-09" "2019-10" "2019-11"
Here we can see the list of months with data for the site ABBY, which is the Abby Road forest in Washington state.
The section $data$siteCodes$availableDataUrls provides the exact API
calls we need in order to query the data for each available site and month.
# Get complete list of available data URLs
wood.urls <- unlist(avail$data$siteCodes$availableDataUrls)
# Total number of URLs
length(wood.urls)
## [1] 535
# Show first 10 URLs available
wood.urls[1:10]
## [1] "https://data.neonscience.org/api/v0/data/DP1.10098.001/ABBY/2015-07"
## [2] "https://data.neonscience.org/api/v0/data/DP1.10098.001/ABBY/2015-08"
## [3] "https://data.neonscience.org/api/v0/data/DP1.10098.001/ABBY/2016-08"
## [4] "https://data.neonscience.org/api/v0/data/DP1.10098.001/ABBY/2016-09"
## [5] "https://data.neonscience.org/api/v0/data/DP1.10098.001/ABBY/2016-10"
## [6] "https://data.neonscience.org/api/v0/data/DP1.10098.001/ABBY/2016-11"
## [7] "https://data.neonscience.org/api/v0/data/DP1.10098.001/ABBY/2017-03"
## [8] "https://data.neonscience.org/api/v0/data/DP1.10098.001/ABBY/2017-04"
## [9] "https://data.neonscience.org/api/v0/data/DP1.10098.001/ABBY/2017-07"
## [10] "https://data.neonscience.org/api/v0/data/DP1.10098.001/ABBY/2017-08"
These URLs are the API calls we can use to find out what files are available for each month where there are data. They are pre-constructed calls to the /data endpoint of the NEON API.
Let's look at the woody plant data from the Rocky Mountain National Park
(RMNP) site from October 2019. We can do this by using the GET() function
on the relevant URL, which we can extract using the grep() function.
Note that if you want data from more than one site/month you need to iterate
this code, GET() fails if you give it more than one URL at a time.
# Get available data for RMNP Oct 2019
woody <- GET(wood.urls[grep("RMNP/2019-10", wood.urls)])
woody.files <- jsonlite::fromJSON(content(woody, as="text"))
# See what files are available for this site and month
woody.files$data$files$name
## [1] "NEON.D10.RMNP.DP1.10098.001.EML.20191010-20191017.20210123T023002Z.xml"
## [2] "NEON.D10.RMNP.DP1.10098.001.2019-10.basic.20210114T173951Z.zip"
## [3] "NEON.D10.RMNP.DP1.10098.001.vst_apparentindividual.2019-10.basic.20210114T173951Z.csv"
## [4] "NEON.D10.RMNP.DP1.10098.001.variables.20210114T173951Z.csv"
## [5] "NEON.D10.RMNP.DP0.10098.001.categoricalCodes.20210114T173951Z.csv"
## [6] "NEON.D10.RMNP.DP1.10098.001.readme.20210123T023002Z.txt"
## [7] "NEON.D10.RMNP.DP1.10098.001.vst_perplotperyear.2019-10.basic.20210114T173951Z.csv"
## [8] "NEON.D10.RMNP.DP1.10098.001.vst_mappingandtagging.basic.20210114T173951Z.csv"
## [9] "NEON.D10.RMNP.DP0.10098.001.validation.20210114T173951Z.csv"
If you've downloaded NEON data before via the data portal or the
neonUtilities package, this should look very familiar. The format
for most of the file names is:
NEON.[domain number].[site code].[data product ID].[file-specific name]. [year and month of data].[basic or expanded data package]. [date of file creation]
Some files omit the year and month, and/or the data package, since they're not specific to a particular measurement interval, such as the data product readme and variables files. The date of file creation uses the ISO6801 format, for example 20210114T173951Z, and can be used to determine whether data have been updated since the last time you downloaded.
Available files in our query for October 2019 at Rocky Mountain are all of the following (leaving off the initial NEON.D10.RMNP.DP1.10098.001):
-
~.vst_perplotperyear.2019-10.basic.20210114T173951Z.csv: data table of measurements conducted at the plot level every year
-
~.vst_apparentindividual.2019-10.basic.20210114T173951Z.csv: data table containing measurements and observations conducted on woody individuals
-
~.vst_mappingandtagging.basic.20210114T173951Z.csv: data table containing mapping data for each measured woody individual. Note year and month are not in file name; these data are collected once per individual and provided with every month of data downloaded
-
~.categoricalCodes.20210114T173951Z.csv: definitions of the values in categorical variables
-
~.readme.20210123T023002Z.txt: readme for the data product (not specific to dates or location)
-
~.EML.20191010-20191017.20210123T023002Z.xml: Ecological Metadata Language (EML) file, describing the data product
-
~.validation.20210114T173951Z.csv: validation file for the data product, lists input data and data entry validation rules
-
~.variables.20210114T173951Z.csv: variables file for the data product, lists data fields in downloaded tables
-
~.2019-10.basic.20210114T173951Z.zip: zip of all files in the basic package. Pre-packaged zips are planned to be removed; may not appear in response to your query
This data product doesn't have an expanded package, so we only see the basic package data files, and only one copy of each of the metadata files.
Let's get the data table for the mapping and tagging data. The list of files doesn't return in the same order every time, so we shouldn't use the position in the list to select the file name we want. Plus, we want code we can re-use when getting data from other sites and other months. So we select file urls based on the data table name in the file names.
vst.maptag <- read.csv(woody.files$data$files$url
[grep("mappingandtagging",
woody.files$data$files$name)])
Note that if there were an expanded package, the code above would return two URLs. In that case you would need to specify the package as well in selecting the URL.
Now we have the data and can access it in R. Just to show that the file we pulled has actual data in it, let's make a quick graphic of the species present and their abundances:
# Get counts by species
countBySp <- table(vst.maptag$taxonID)
# Reorder so list is ordered most to least abundance
countBySp <- countBySp[order(countBySp, decreasing=T)]
# Plot abundances
barplot(countBySp, names.arg=names(countBySp),
ylab="Total", las=2)
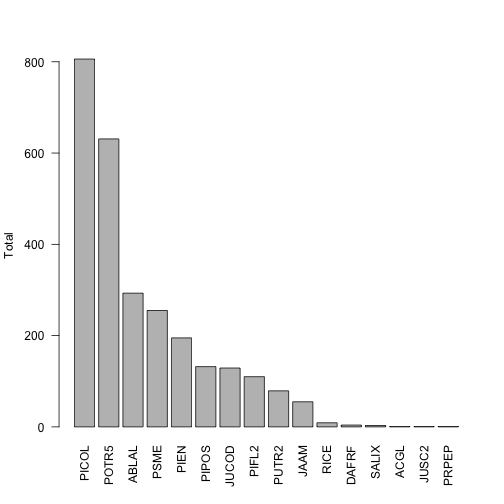
This shows us that the two most abundant species are designated with the taxon codes PICOL and POTR5. We can look back at the data table, check the scientificName field corresponding to these values, and see that these are lodgepole pine and quaking aspen, as we might expect in the eastern foothills of the Rocky mountains.
Let's say we're interested in how NEON defines quaking aspen, and what taxon authority it uses for its definition. We can use the /taxonomy endpoint of the API to do that.
Taxonomy
NEON maintains accepted taxonomies for many of the taxonomic identification data we collect. NEON taxonomies are available for query via the API; they are also provided via an interactive user interface, the Taxon Viewer.
NEON taxonomy data provides the reference information for how NEON validates taxa; an identification must appear in the taxonomy lists in order to be accepted into the NEON database. Additions to the lists are reviewed regularly. The taxonomy lists also provide the author of the scientific name, and the reference text used.
The taxonomy endpoint of the API has query options that are a bit more complicated than what was described in the "Anatomy of an API Call" section above. As described above, each endpoint has a single type of target - a data product number, a named location name, etc. For taxonomic data, there are multiple query options, and some of them can be used in combination. Instead of entering a single target, we specify the query type, and then the query parameter to search for. For example, a query for taxa in the Pinaceae family:
http://data.neonscience.org/api/v0/taxonomy/?family=Pinaceae
The available types of queries are listed in the taxonomy section of the API web page. Briefly, they are:
-
taxonTypeCode: Which of the taxonomies maintained by NEON are you looking for? BIRD, FISH, PLANT, etc. Cannot be used in combination with the taxonomic rank queries. - each of the major taxonomic ranks from genus through kingdom
-
scientificname: Genus + specific epithet (+ authority). Search is by exact match only, see final example below. -
verbose: Do you want the short (false) or long (true) response -
offset: Skip this number of items at the start of the list. -
limit: Result set will be truncated at this length.
For the first example, let's query for the loon family, Gaviidae, in the bird taxonomy. Note that query parameters are case-sensitive.
loon.req <- GET("http://data.neonscience.org/api/v0/taxonomy/?family=Gaviidae")
Parse the results into a list using fromJSON():
loon.list <- jsonlite::fromJSON(content(loon.req, as="text"))
And look at the $data element of the results, which contains:
- The full taxonomy of each taxon
- The short taxon code used by NEON (taxonID/acceptedTaxonID)
- The author of the scientific name (scientificNameAuthorship)
- The vernacular name, if applicable
- The reference text used (nameAccordingToID)
The terms used for each field are matched to Darwin Core (dwc) and the Global Biodiversity Information Facility (gbif) terms, where possible, and the matches are indicated in the column headers.
loon.list$data
## taxonTypeCode taxonID acceptedTaxonID dwc:scientificName dwc:scientificNameAuthorship dwc:taxonRank
## 1 BIRD ARLO ARLO Gavia arctica (Linnaeus) species
## 2 BIRD COLO COLO Gavia immer (Brunnich) species
## 3 BIRD PALO PALO Gavia pacifica (Lawrence) species
## 4 BIRD RTLO RTLO Gavia stellata (Pontoppidan) species
## 5 BIRD YBLO YBLO Gavia adamsii (G. R. Gray) species
## dwc:vernacularName dwc:nameAccordingToID dwc:kingdom dwc:phylum dwc:class dwc:order dwc:family
## 1 Arctic Loon doi: 10.1642/AUK-15-73.1 Animalia Chordata Aves Gaviiformes Gaviidae
## 2 Common Loon doi: 10.1642/AUK-15-73.1 Animalia Chordata Aves Gaviiformes Gaviidae
## 3 Pacific Loon doi: 10.1642/AUK-15-73.1 Animalia Chordata Aves Gaviiformes Gaviidae
## 4 Red-throated Loon doi: 10.1642/AUK-15-73.1 Animalia Chordata Aves Gaviiformes Gaviidae
## 5 Yellow-billed Loon doi: 10.1642/AUK-15-73.1 Animalia Chordata Aves Gaviiformes Gaviidae
## dwc:genus gbif:subspecies gbif:variety
## 1 Gavia NA NA
## 2 Gavia NA NA
## 3 Gavia NA NA
## 4 Gavia NA NA
## 5 Gavia NA NA
To get the entire list for a particular taxonomic type, use the
taxonTypeCode query. Be cautious with this query, the PLANT taxonomic
list has several hundred thousand entries.
For an example, let's look up the small mammal taxonomic list, which
is one of the shorter ones, and use the verbose=true option to see
a more extensive list of taxon data, including many taxon ranks that
aren't populated for these taxa. For space here, we'll display only
the first 10 taxa:
mam.req <- GET("http://data.neonscience.org/api/v0/taxonomy/?taxonTypeCode=SMALL_MAMMAL&verbose=true")
mam.list <- jsonlite::fromJSON(content(mam.req, as="text"))
mam.list$data[1:10,]
## taxonTypeCode taxonID acceptedTaxonID dwc:scientificName dwc:scientificNameAuthorship
## 1 SMALL_MAMMAL AMHA AMHA Ammospermophilus harrisii Audubon and Bachman
## 2 SMALL_MAMMAL AMIN AMIN Ammospermophilus interpres Merriam
## 3 SMALL_MAMMAL AMLE AMLE Ammospermophilus leucurus Merriam
## 4 SMALL_MAMMAL AMLT AMLT Ammospermophilus leucurus tersus Goldman
## 5 SMALL_MAMMAL AMNE AMNE Ammospermophilus nelsoni Merriam
## 6 SMALL_MAMMAL AMSP AMSP Ammospermophilus sp. <NA>
## 7 SMALL_MAMMAL APRN APRN Aplodontia rufa nigra Taylor
## 8 SMALL_MAMMAL APRU APRU Aplodontia rufa Rafinesque
## 9 SMALL_MAMMAL ARAL ARAL Arborimus albipes Merriam
## 10 SMALL_MAMMAL ARLO ARLO Arborimus longicaudus True
## dwc:taxonRank dwc:vernacularName taxonProtocolCategory dwc:nameAccordingToID
## 1 species Harriss Antelope Squirrel opportunistic isbn: 978 0801882210
## 2 species Texas Antelope Squirrel opportunistic isbn: 978 0801882210
## 3 species Whitetailed Antelope Squirrel opportunistic isbn: 978 0801882210
## 4 subspecies <NA> opportunistic isbn: 978 0801882210
## 5 species Nelsons Antelope Squirrel opportunistic isbn: 978 0801882210
## 6 genus <NA> opportunistic isbn: 978 0801882210
## 7 subspecies <NA> non-target isbn: 978 0801882210
## 8 species Sewellel non-target isbn: 978 0801882210
## 9 species Whitefooted Vole target isbn: 978 0801882210
## 10 species Red Tree Vole target isbn: 978 0801882210
## dwc:nameAccordingToTitle
## 1 Wilson D. E. and D. M. Reeder. 2005. Mammal Species of the World; A Taxonomic and Geographic Reference. Third edition. Johns Hopkins University Press; Baltimore, MD.
## 2 Wilson D. E. and D. M. Reeder. 2005. Mammal Species of the World; A Taxonomic and Geographic Reference. Third edition. Johns Hopkins University Press; Baltimore, MD.
## 3 Wilson D. E. and D. M. Reeder. 2005. Mammal Species of the World; A Taxonomic and Geographic Reference. Third edition. Johns Hopkins University Press; Baltimore, MD.
## 4 Wilson D. E. and D. M. Reeder. 2005. Mammal Species of the World; A Taxonomic and Geographic Reference. Third edition. Johns Hopkins University Press; Baltimore, MD.
## 5 Wilson D. E. and D. M. Reeder. 2005. Mammal Species of the World; A Taxonomic and Geographic Reference. Third edition. Johns Hopkins University Press; Baltimore, MD.
## 6 Wilson D. E. and D. M. Reeder. 2005. Mammal Species of the World; A Taxonomic and Geographic Reference. Third edition. Johns Hopkins University Press; Baltimore, MD.
## 7 Wilson D. E. and D. M. Reeder. 2005. Mammal Species of the World; A Taxonomic and Geographic Reference. Third edition. Johns Hopkins University Press; Baltimore, MD.
## 8 Wilson D. E. and D. M. Reeder. 2005. Mammal Species of the World; A Taxonomic and Geographic Reference. Third edition. Johns Hopkins University Press; Baltimore, MD.
## 9 Wilson D. E. and D. M. Reeder. 2005. Mammal Species of the World; A Taxonomic and Geographic Reference. Third edition. Johns Hopkins University Press; Baltimore, MD.
## 10 Wilson D. E. and D. M. Reeder. 2005. Mammal Species of the World; A Taxonomic and Geographic Reference. Third edition. Johns Hopkins University Press; Baltimore, MD.
## dwc:kingdom gbif:subkingdom gbif:infrakingdom gbif:superdivision gbif:division gbif:subdivision
## 1 Animalia NA NA NA NA NA
## 2 Animalia NA NA NA NA NA
## 3 Animalia NA NA NA NA NA
## 4 Animalia NA NA NA NA NA
## 5 Animalia NA NA NA NA NA
## 6 Animalia NA NA NA NA NA
## 7 Animalia NA NA NA NA NA
## 8 Animalia NA NA NA NA NA
## 9 Animalia NA NA NA NA NA
## 10 Animalia NA NA NA NA NA
## gbif:infradivision gbif:parvdivision gbif:superphylum dwc:phylum gbif:subphylum gbif:infraphylum
## 1 NA NA NA Chordata NA NA
## 2 NA NA NA Chordata NA NA
## 3 NA NA NA Chordata NA NA
## 4 NA NA NA Chordata NA NA
## 5 NA NA NA Chordata NA NA
## 6 NA NA NA Chordata NA NA
## 7 NA NA NA Chordata NA NA
## 8 NA NA NA Chordata NA NA
## 9 NA NA NA Chordata NA NA
## 10 NA NA NA Chordata NA NA
## gbif:superclass dwc:class gbif:subclass gbif:infraclass gbif:superorder dwc:order gbif:suborder
## 1 NA Mammalia NA NA NA Rodentia NA
## 2 NA Mammalia NA NA NA Rodentia NA
## 3 NA Mammalia NA NA NA Rodentia NA
## 4 NA Mammalia NA NA NA Rodentia NA
## 5 NA Mammalia NA NA NA Rodentia NA
## 6 NA Mammalia NA NA NA Rodentia NA
## 7 NA Mammalia NA NA NA Rodentia NA
## 8 NA Mammalia NA NA NA Rodentia NA
## 9 NA Mammalia NA NA NA Rodentia NA
## 10 NA Mammalia NA NA NA Rodentia NA
## gbif:infraorder gbif:section gbif:subsection gbif:superfamily dwc:family gbif:subfamily gbif:tribe
## 1 NA NA NA NA Sciuridae Xerinae Marmotini
## 2 NA NA NA NA Sciuridae Xerinae Marmotini
## 3 NA NA NA NA Sciuridae Xerinae Marmotini
## 4 NA NA NA NA Sciuridae Xerinae Marmotini
## 5 NA NA NA NA Sciuridae Xerinae Marmotini
## 6 NA NA NA NA Sciuridae Xerinae Marmotini
## 7 NA NA NA NA Aplodontiidae <NA> <NA>
## 8 NA NA NA NA Aplodontiidae <NA> <NA>
## 9 NA NA NA NA Cricetidae Arvicolinae <NA>
## 10 NA NA NA NA Cricetidae Arvicolinae <NA>
## gbif:subtribe dwc:genus dwc:subgenus gbif:subspecies gbif:variety gbif:subvariety gbif:form
## 1 NA Ammospermophilus <NA> NA NA NA NA
## 2 NA Ammospermophilus <NA> NA NA NA NA
## 3 NA Ammospermophilus <NA> NA NA NA NA
## 4 NA Ammospermophilus <NA> NA NA NA NA
## 5 NA Ammospermophilus <NA> NA NA NA NA
## 6 NA Ammospermophilus <NA> NA NA NA NA
## 7 NA Aplodontia <NA> NA NA NA NA
## 8 NA Aplodontia <NA> NA NA NA NA
## 9 NA Arborimus <NA> NA NA NA NA
## 10 NA Arborimus <NA> NA NA NA NA
## gbif:subform speciesGroup dwc:specificEpithet dwc:infraspecificEpithet
## 1 NA <NA> harrisii <NA>
## 2 NA <NA> interpres <NA>
## 3 NA <NA> leucurus <NA>
## 4 NA <NA> leucurus tersus
## 5 NA <NA> nelsoni <NA>
## 6 NA <NA> sp. <NA>
## 7 NA <NA> rufa nigra
## 8 NA <NA> rufa <NA>
## 9 NA <NA> albipes <NA>
## 10 NA <NA> longicaudus <NA>
Now let's go back to our question about quaking aspen. To get
information about a single taxon, use the scientificname
query. This query will not do a fuzzy match, so you need to query
the exact name of the taxon in the NEON taxonomy. Because of this,
the query will be most useful in cases like the current one, where
you already have NEON data in hand and are looking for more
information about a specific taxon. Querying on scientificname
is unlikely to be an efficient way to figure out if NEON recognizes
a particular taxon.
In addition, scientific names contain spaces, which are not allowed in a URL. The spaces need to be replaced with the URL encoding replacement, %20.
Looking up the POTR5 data in the woody vegetation product, we
see that the scientific name is Populus tremuloides Michx.
This means we need to search for Populus%20tremuloides%20Michx.
to get the exact match.
aspen.req <- GET("http://data.neonscience.org/api/v0/taxonomy/?scientificname=Populus%20tremuloides%20Michx.")
aspen.list <- jsonlite::fromJSON(content(aspen.req, as="text"))
aspen.list$data
## taxonTypeCode taxonID acceptedTaxonID dwc:scientificName dwc:scientificNameAuthorship
## 1 PLANT POTR5 POTR5 Populus tremuloides Michx. Michx.
## dwc:taxonRank dwc:vernacularName dwc:nameAccordingToID dwc:kingdom dwc:phylum
## 1 species quaking aspen http://plants.usda.gov (accessed 8/25/2014) Plantae Magnoliophyta
## dwc:class dwc:order dwc:family dwc:genus gbif:subspecies gbif:variety
## 1 Magnoliopsida Salicales Salicaceae Populus NA NA
This shows us the definition for Populus tremuloides Michx. does
not include a subspecies or variety, and the authority for the
taxon information (nameAccordingToID) is the USDA PLANTS
database. This means NEON taxonomic definitions are aligned with
the USDA, and is true for the large majority of plants in the
NEON taxon system.
Spatial data
How to get spatial data and what to do with it depends on which type of data you're working with.
Instrumentation data (both aquatic and terrestrial)
The sensor_positions files, which are included in the list of available files, contain spatial coordinates for each sensor in the data. See the final section of the Geolocation tutorial for guidance in using these files.
Observational data - Aquatic
Latitude, longitude, elevation, and associated uncertainties are included in data downloads. Most products also include an "additional coordinate uncertainty" that should be added to the provided uncertainty. Additional spatial data, such as northing and easting, can be downloaded from the API.
Observational data - Terrestrial
Latitude, longitude, elevation, and associated uncertainties are included in
data downloads. These are the coordinates and uncertainty of the sampling plot;
for many protocols it is possible to calculate a more precise location.
Instructions for doing this are in the respective data product user guides, and
code is in the geoNEON package on GitHub.
Querying a single named location
Let's look at a specific sampling location in the woody vegetation structure
data we downloaded above. To do this, look for the field called namedLocation,
which is present in all observational data products, both aquatic and
terrestrial. This field matches the exact name of the location in the NEON
database.
head(vst.maptag$namedLocation)
## [1] "RMNP_043.basePlot.vst" "RMNP_043.basePlot.vst" "RMNP_043.basePlot.vst" "RMNP_043.basePlot.vst"
## [5] "RMNP_043.basePlot.vst" "RMNP_043.basePlot.vst"
Here we see the first six entries in the namedLocation column, which tells us
the names of the Terrestrial Observation plots where the woody plant surveys
were conducted.
We can query the locations endpoint of the API for the first named location,
RMNP_043.basePlot.vst.
req.loc <- GET("http://data.neonscience.org/api/v0/locations/RMNP_043.basePlot.vst")
vst.RMNP_043 <- jsonlite::fromJSON(content(req.loc, as="text"))
vst.RMNP_043
## $data
## $data$locationName
## [1] "RMNP_043.basePlot.vst"
##
## $data$locationDescription
## [1] "Plot \"RMNP_043\" at site \"RMNP\""
##
## $data$locationType
## [1] "OS Plot - vst"
##
## $data$domainCode
## [1] "D10"
##
## $data$siteCode
## [1] "RMNP"
##
## $data$locationDecimalLatitude
## [1] 40.27683
##
## $data$locationDecimalLongitude
## [1] -105.5454
##
## $data$locationElevation
## [1] 2740.39
##
## $data$locationUtmEasting
## [1] 453634.6
##
## $data$locationUtmNorthing
## [1] 4458626
##
## $data$locationUtmHemisphere
## [1] "N"
##
## $data$locationUtmZone
## [1] 13
##
## $data$alphaOrientation
## [1] 0
##
## $data$betaOrientation
## [1] 0
##
## $data$gammaOrientation
## [1] 0
##
## $data$xOffset
## [1] 0
##
## $data$yOffset
## [1] 0
##
## $data$zOffset
## [1] 0
##
## $data$offsetLocation
## NULL
##
## $data$locationProperties
## locationPropertyName locationPropertyValue
## 1 Value for Coordinate source GeoXH 6000
## 2 Value for Coordinate uncertainty 0.09
## 3 Value for Country unitedStates
## 4 Value for County Larimer
## 5 Value for Elevation uncertainty 0.1
## 6 Value for Filtered positions 300
## 7 Value for Geodetic datum WGS84
## 8 Value for Horizontal dilution of precision 1
## 9 Value for Maximum elevation 2743.43
## 10 Value for Minimum elevation 2738.52
## 11 Value for National Land Cover Database (2001) evergreenForest
## 12 Value for Plot dimensions 40m x 40m
## 13 Value for Plot ID RMNP_043
## 14 Value for Plot size 1600
## 15 Value for Plot subtype basePlot
## 16 Value for Plot type tower
## 17 Value for Positional dilution of precision 1.8
## 18 Value for Reference Point Position 41
## 19 Value for Slope aspect 0
## 20 Value for Slope gradient 0
## 21 Value for Soil type order Alfisols
## 22 Value for State province CO
## 23 Value for UTM Zone 13N
##
## $data$locationParent
## [1] "RMNP"
##
## $data$locationParentUrl
## [1] "https://data.neonscience.org/api/v0/locations/RMNP"
##
## $data$locationChildren
## [1] "RMNP_043.basePlot.vst.41" "RMNP_043.basePlot.vst.43" "RMNP_043.basePlot.vst.49"
## [4] "RMNP_043.basePlot.vst.51" "RMNP_043.basePlot.vst.59" "RMNP_043.basePlot.vst.57"
## [7] "RMNP_043.basePlot.vst.25" "RMNP_043.basePlot.vst.21" "RMNP_043.basePlot.vst.23"
## [10] "RMNP_043.basePlot.vst.33" "RMNP_043.basePlot.vst.31" "RMNP_043.basePlot.vst.39"
## [13] "RMNP_043.basePlot.vst.61"
##
## $data$locationChildrenUrls
## [1] "https://data.neonscience.org/api/v0/locations/RMNP_043.basePlot.vst.41"
## [2] "https://data.neonscience.org/api/v0/locations/RMNP_043.basePlot.vst.43"
## [3] "https://data.neonscience.org/api/v0/locations/RMNP_043.basePlot.vst.49"
## [4] "https://data.neonscience.org/api/v0/locations/RMNP_043.basePlot.vst.51"
## [5] "https://data.neonscience.org/api/v0/locations/RMNP_043.basePlot.vst.59"
## [6] "https://data.neonscience.org/api/v0/locations/RMNP_043.basePlot.vst.57"
## [7] "https://data.neonscience.org/api/v0/locations/RMNP_043.basePlot.vst.25"
## [8] "https://data.neonscience.org/api/v0/locations/RMNP_043.basePlot.vst.21"
## [9] "https://data.neonscience.org/api/v0/locations/RMNP_043.basePlot.vst.23"
## [10] "https://data.neonscience.org/api/v0/locations/RMNP_043.basePlot.vst.33"
## [11] "https://data.neonscience.org/api/v0/locations/RMNP_043.basePlot.vst.31"
## [12] "https://data.neonscience.org/api/v0/locations/RMNP_043.basePlot.vst.39"
## [13] "https://data.neonscience.org/api/v0/locations/RMNP_043.basePlot.vst.61"
Note spatial information under $data$[nameOfCoordinate] and under
$data$locationProperties. These coordinates represent the centroid of
plot RMNP_043, and should match the coordinates for the plot in the
vst_perplotperyear data table. In rare cases, spatial data may be
updated, and if this has happened more recently than the data table
was published, there may be a small mismatch in the coordinates. In
those cases, the data accessed via the API will be the most up to date.
Also note $data$locationChildren: these are the
point and subplot locations within plot RMNP_043. And
$data$locationChildrenUrls provides pre-constructed API calls for
querying those child locations. Let's look up the location data for
point 31 in plot RMNP_043.
req.child.loc <- GET(grep("31",
vst.RMNP_043$data$locationChildrenUrls,
value=T))
vst.RMNP_043.31 <- jsonlite::fromJSON(content(req.child.loc, as="text"))
vst.RMNP_043.31
## $data
## $data$locationName
## [1] "RMNP_043.basePlot.vst.31"
##
## $data$locationDescription
## [1] "31"
##
## $data$locationType
## [1] "Point"
##
## $data$domainCode
## [1] "D10"
##
## $data$siteCode
## [1] "RMNP"
##
## $data$locationDecimalLatitude
## [1] 40.27674
##
## $data$locationDecimalLongitude
## [1] -105.5455
##
## $data$locationElevation
## [1] 2741.56
##
## $data$locationUtmEasting
## [1] 453623.7
##
## $data$locationUtmNorthing
## [1] 4458616
##
## $data$locationUtmHemisphere
## [1] "N"
##
## $data$locationUtmZone
## [1] 13
##
## $data$alphaOrientation
## [1] 0
##
## $data$betaOrientation
## [1] 0
##
## $data$gammaOrientation
## [1] 0
##
## $data$xOffset
## [1] 0
##
## $data$yOffset
## [1] 0
##
## $data$zOffset
## [1] 0
##
## $data$offsetLocation
## NULL
##
## $data$locationProperties
## locationPropertyName locationPropertyValue
## 1 Value for Coordinate source GeoXH 6000
## 2 Value for Coordinate uncertainty 0.16
## 3 Value for Elevation uncertainty 0.28
## 4 Value for Geodetic datum WGS84
## 5 Value for National Land Cover Database (2001) evergreenForest
## 6 Value for Point ID 31
## 7 Value for UTM Zone 13N
##
## $data$locationParent
## [1] "RMNP_043.basePlot.vst"
##
## $data$locationParentUrl
## [1] "https://data.neonscience.org/api/v0/locations/RMNP_043.basePlot.vst"
##
## $data$locationChildren
## list()
##
## $data$locationChildrenUrls
## list()
Looking at the easting and northing coordinates, we can see that point 31 is about 10 meters away from the plot centroid in both directions. Point 31 has no child locations.
For the records with pointID=31 in the vst.maptag table we
downloaded, these coordinates are the reference location that could be
used together with the stemDistance and stemAzimuth fields to
calculate the precise locations of individual trees. For detailed
instructions in how to do this, see the data product user guide.
Alternatively, the geoNEON package contains functions to make this
calculation for you, including accessing the location data from the
API. See below for details and links to tutorials.
R Packages
NEON provides two customized R packages that can carry out many of the operations described above, in addition to other data transformations.
neonUtilities
The neonUtilities package contains functions that download data
via the API, and that merge the individual files for each site and
month in a single continuous file for each type of data table in the
download.
For a guide to using neonUtilities to download and stack data files,
check out the Download and Explore tutorial.
geoNEON
The geoNEON package contains functions that access spatial data
via the API, and that calculate precise locations for terrestrial
observational data. As in the case of woody vegetation structure,
terrestrial observational data products are published with
spatial data at the plot level, but more precise sampling locations
are usually possible to calculate.
For a guide to using geoNEON to calculate sampling locations,
check out the Geolocation tutorial.
R
In this workshop we will primarily be working with Microsoft Excel to view and work with NEON data. If you are interested in learning more about the R "ecosystem", consider reading Mark Seller's A Field Guide to the R Ecosystem. This is not a how-to set of materials but rather, in his words, a "field guide to introduce the reader to the main components of the R ecosystem that may be encountered in “the field”."
R Tutorials for Working with NEON Data
During the working with NEON data products portion of the workshop we will primarily be working in spreadsheets. The tutorials below are a few of the available ones that show you have to do similar things with NEON data in R.