Series
Introduction to Working With Time Series Data in Text Formats in R
The tutorials in this series cover how to open, work with and plot tabular time-series data in R. Additional topics include working with time and date classes (e.g., POSIXct, POSIXlt, and Date), subsetting time series data by date and time and created facetted or tiles sets of plots.
Data used in this series cover NEON Harvard Forest Field Site and are in .csv file format.
Series Objectives
After completing the series you will:
-
Time Series 00
- Be able to open a
.csvfile inRusingread.csv()and understand why we are using that file type. - Understand how to work data stored in different columns within a
data.frameinR. - Understand how to examine
Robject structures and dataclasses. - Be able to convert dates, stored as a character class, into an
Rdate class. - Know how to create a quick plot of a time-series data set using
qplot.
- Be able to open a
-
Time Series 01
- Know how to import a .csv file and examine the structure of the related
Robject. - Use a metadata file to better understand the content of a dataset.
- Understand the importance of including metadata details in your
Rscript. - Know what an EML file is.
- Know how to import a .csv file and examine the structure of the related
-
Time Series 02
- Understand various date-time classes and data structure in
R. - Understand what
POSIXctandPOSIXltdata classes are and why POSIXct may be preferred for some tasks. - Be able to convert a column containing date-time information in character
format to a date-time
Rclass. - Be able to convert a date-time column to different date-time classes.
- Learn how to write out a date-time class object in different ways (month-day, month-day-year, etc).
- Understand various date-time classes and data structure in
-
Time Series 03
- Be able to subset data by date.
- Know how to search for NA or missing data values.
- Understand different possibilities on how to deal with missing data.
-
Time Series 04
- Know several ways to manipulate data using functions in the
dplyrpackage inR. - Be able to use
group-by(),summarize(), andmutate()functions. - Write and understand
Rcode with pipes for cleaner, efficient coding. - Use the
year()function from thelubridatepackage to extract year from a date-time class variable.
- Know several ways to manipulate data using functions in the
-
Time Series 05
- Be able to create basic time series plots using
ggplot()inR. - Understand the syntax of
ggplot()and know how to find out more about the package. - Be able to plot data using scatter and bar plots.
- Be able to create basic time series plots using
-
Time Series 06
- Know how to use
facets()in theggplot2package. - Be able to combine different types of data into one plot layout.
- Know how to use
-
Time Series Culmination Activity
- have applied
ggplot2anddplyrskills to a new data set. - learn how to set min/max axis values in
ggplot()to align data on multiple plots.
- have applied
Things You’ll Need To Complete This Series
Setup RStudio
To complete the tutorial series you will need an updated version of R and,
preferably, RStudio installed on your computer.
R
is a programming language that specializes in statistical computing. It is a
powerful tool for exploratory data analysis. To interact with R, we strongly
recommend
RStudio,
an interactive development environment (IDE).
Install R Packages
You can chose to install packages with each lesson or you can download all
of the necessary R packages now.
-
ggplot2:
install.packages("ggplot2") -
lubridate:
install.packages("lubridate") -
dplyr:
install.packages("dplyr") -
scales:
install.packages("scales") -
gridExtra:
install.packages("gridExtra") -
ggthemes:
install.packages("ggthemes") -
reshape2:
install.packages("reshape2") -
zoo:
install.packages("zoo")
More on Packages in R - Adapted from Software Carpentry.
Working with Spatio-temporal Data in R Series: This tutorial series is part of a larger spatio-temporal tutorial series and Data Carpentry workshop. Included series are working with raster data in R, working with vector data in R and working with tabular time series in R.
Time Series 00: Intro to Time Series Data in R - Managing Date/Time Formats
Authors: Megan A. Jones, Marisa Guarinello, Courtney Soderberg, Leah A. Wasser
Last Updated: May 13, 2021
This tutorial will demonstrate how to import a time series dataset stored in .csv
format into R. It will explore data classes for columns in a data.frame and
will walk through how to
convert a date, stored as a character string, into a date class that R can
recognize and plot efficiently.
Learning Objectives
After completing this tutorial, you will be able to:
- Open a
.csvfile in R usingread.csv()and understand why we are using that file type. - Work with data stored in different columns within a
data.framein R. - Examine R object structures and data
classes. - Convert dates, stored as a character class, into an R date class.
- Create a quick plot of a time-series dataset using
qplot.
Things You’ll Need To Complete This Tutorial
You will need the most current version of R and, preferably, RStudio loaded on your computer to complete this tutorial.
Install R Packages
-
ggplot2:
install.packages("ggplot2")
More on Packages in R – Adapted from Software Carpentry.
Download Data
NEON Teaching Data Subset: Meteorological Data for Harvard Forest
The data used in this lesson were collected at the National Ecological Observatory Network's Harvard Forest field site. These data are proxy data for what will be available for 30 years on the NEON data portal for the Harvard Forest and other field sites located across the United States.
Set Working Directory: This lesson assumes that you have set your working directory to the location of the downloaded and unzipped data subsets.
An overview of setting the working directory in R can be found here.
R Script & Challenge Code: NEON data lessons often contain challenges that reinforce learned skills. If available, the code for challenge solutions is found in the downloadable R script of the entire lesson, available in the footer of each lesson page.
Data Related to Phenology
In this tutorial, we will explore atmospheric data (including temperature, precipitation and other metrics) collected by sensors mounted on a flux tower at the NEON Harvard Forest field site. We are interested in exploring changes in temperature, precipitation, Photosynthetically Active Radiation (PAR) and day length throughout the year -- metrics that impact changes in the timing of plant phenophases (phenology).
About .csv Format
The data that we will use is in .csv (comma-separated values) file format. The
.csv format is a plain text format, where each value in the dataset is
separate by a comma and each "row" in the dataset is separated by a line break.
Plain text formats are ideal for working both across platforms (Mac, PC, LINUX,
etc) and also can be read by many different tools. The plain text
format is also less likely to become obsolete over time.
Import the Data
To begin, let's import the data into R. We can use base R functionality
to import a .csv file. We will use the ggplot2 package to plot our data.
# Load packages required for entire script.
# library(PackageName) # purpose of package
library(ggplot2) # efficient, pretty plotting - required for qplot function
# set working directory to ensure R can find the file we wish to import
# provide the location for where you've unzipped the lesson data
wd <- "~/Git/data/"
Once our working directory is set, we can import the file using read.csv().
# Load csv file of daily meteorological data from Harvard Forest
harMet.daily <- read.csv(
file=paste0(wd,"NEON-DS-Met-Time-Series/HARV/FisherTower-Met/hf001-06-daily-m.csv"),
stringsAsFactors = FALSE
)
stringsAsFactors=FALSE
When reading in files we most often use stringsAsFactors = FALSE. This
setting ensures that non-numeric data (strings) are not converted to
factors.
What Is A Factor?
A factor is similar to a category. However factors can be numerically interpreted (they can have an order) and may have a level associated with them.
Examples of factors:
- Month Names (an ordinal variable): Month names are non-numerical but we know that April (month 4) comes after March (month 3) and each could be represented by a number (4 & 3).
- 1 and 2s to represent male and female sex (a nominal variable): Numerical interpretation of non-numerical data but no order to the levels.
After loading the data it is easy to convert any field that should be a factor by
using as.factor(). Therefore it is often best to read in a file with
stringsAsFactors = FALSE.
Data.Frames in R
The read.csv() imports our .csv into a data.frame object in R. data.frames
are ideal for working with tabular data - they are similar to a spreadsheet.
# what type of R object is our imported data?
class(harMet.daily)
## [1] "data.frame"
Data Structure
Once the data are imported, we can explore their structure. There are several ways to examine the structure of a data frame:
-
head(): shows us the first 6 rows of the data (tail()shows the last 6 rows). -
str(): displays the structure of the data as R interprets it.
Let's use both to explore our data.
# view first 6 rows of the dataframe
head(harMet.daily)
## date jd airt f.airt airtmax f.airtmax airtmin f.airtmin rh
## 1 2001-02-11 42 -10.7 -6.9 -15.1 40
## 2 2001-02-12 43 -9.8 -2.4 -17.4 45
## 3 2001-02-13 44 -2.0 5.7 -7.3 70
## 4 2001-02-14 45 -0.5 1.9 -5.7 78
## 5 2001-02-15 46 -0.4 2.4 -5.7 69
## 6 2001-02-16 47 -3.0 1.3 -9.0 82
## f.rh rhmax f.rhmax rhmin f.rhmin dewp f.dewp dewpmax f.dewpmax
## 1 58 22 -22.2 -16.8
## 2 85 14 -20.7 -9.2
## 3 100 34 -7.6 -4.6
## 4 100 59 -4.1 1.9
## 5 100 37 -6.0 2.0
## 6 100 46 -5.9 -0.4
## dewpmin f.dewpmin prec f.prec slrt f.slrt part f.part netr f.netr
## 1 -25.7 0.0 14.9 NA M NA M
## 2 -27.9 0.0 14.8 NA M NA M
## 3 -10.2 0.0 14.8 NA M NA M
## 4 -10.2 6.9 2.6 NA M NA M
## 5 -12.1 0.0 10.5 NA M NA M
## 6 -10.6 2.3 6.4 NA M NA M
## bar f.bar wspd f.wspd wres f.wres wdir f.wdir wdev f.wdev gspd
## 1 1025 3.3 2.9 287 27 15.4
## 2 1033 1.7 0.9 245 55 7.2
## 3 1024 1.7 0.9 278 53 9.6
## 4 1016 2.5 1.9 197 38 11.2
## 5 1010 1.6 1.2 300 40 12.7
## 6 1016 1.1 0.5 182 56 5.8
## f.gspd s10t f.s10t s10tmax f.s10tmax s10tmin f.s10tmin
## 1 NA M NA M NA M
## 2 NA M NA M NA M
## 3 NA M NA M NA M
## 4 NA M NA M NA M
## 5 NA M NA M NA M
## 6 NA M NA M NA M
# View the structure (str) of the data
str(harMet.daily)
## 'data.frame': 5345 obs. of 46 variables:
## $ date : chr "2001-02-11" "2001-02-12" "2001-02-13" "2001-02-14" ...
## $ jd : int 42 43 44 45 46 47 48 49 50 51 ...
## $ airt : num -10.7 -9.8 -2 -0.5 -0.4 -3 -4.5 -9.9 -4.5 3.2 ...
## $ f.airt : chr "" "" "" "" ...
## $ airtmax : num -6.9 -2.4 5.7 1.9 2.4 1.3 -0.7 -3.3 0.7 8.9 ...
## $ f.airtmax: chr "" "" "" "" ...
## $ airtmin : num -15.1 -17.4 -7.3 -5.7 -5.7 -9 -12.7 -17.1 -11.7 -1.3 ...
## $ f.airtmin: chr "" "" "" "" ...
## $ rh : int 40 45 70 78 69 82 66 51 57 62 ...
## $ f.rh : chr "" "" "" "" ...
## $ rhmax : int 58 85 100 100 100 100 100 71 81 78 ...
## $ f.rhmax : chr "" "" "" "" ...
## $ rhmin : int 22 14 34 59 37 46 30 34 37 42 ...
## $ f.rhmin : chr "" "" "" "" ...
## $ dewp : num -22.2 -20.7 -7.6 -4.1 -6 -5.9 -10.8 -18.5 -12 -3.5 ...
## $ f.dewp : chr "" "" "" "" ...
## $ dewpmax : num -16.8 -9.2 -4.6 1.9 2 -0.4 -0.7 -14.4 -4 0.6 ...
## $ f.dewpmax: chr "" "" "" "" ...
## $ dewpmin : num -25.7 -27.9 -10.2 -10.2 -12.1 -10.6 -25.4 -25 -16.5 -5.7 ...
## $ f.dewpmin: chr "" "" "" "" ...
## $ prec : num 0 0 0 6.9 0 2.3 0 0 0 0 ...
## $ f.prec : chr "" "" "" "" ...
## $ slrt : num 14.9 14.8 14.8 2.6 10.5 6.4 10.3 15.5 15 7.7 ...
## $ f.slrt : chr "" "" "" "" ...
## $ part : num NA NA NA NA NA NA NA NA NA NA ...
## $ f.part : chr "M" "M" "M" "M" ...
## $ netr : num NA NA NA NA NA NA NA NA NA NA ...
## $ f.netr : chr "M" "M" "M" "M" ...
## $ bar : int 1025 1033 1024 1016 1010 1016 1008 1022 1022 1017 ...
## $ f.bar : chr "" "" "" "" ...
## $ wspd : num 3.3 1.7 1.7 2.5 1.6 1.1 3.3 2 2.5 2 ...
## $ f.wspd : chr "" "" "" "" ...
## $ wres : num 2.9 0.9 0.9 1.9 1.2 0.5 3 1.9 2.1 1.8 ...
## $ f.wres : chr "" "" "" "" ...
## $ wdir : int 287 245 278 197 300 182 281 272 217 218 ...
## $ f.wdir : chr "" "" "" "" ...
## $ wdev : int 27 55 53 38 40 56 24 24 31 27 ...
## $ f.wdev : chr "" "" "" "" ...
## $ gspd : num 15.4 7.2 9.6 11.2 12.7 5.8 16.9 10.3 11.1 10.9 ...
## $ f.gspd : chr "" "" "" "" ...
## $ s10t : num NA NA NA NA NA NA NA NA NA NA ...
## $ f.s10t : chr "M" "M" "M" "M" ...
## $ s10tmax : num NA NA NA NA NA NA NA NA NA NA ...
## $ f.s10tmax: chr "M" "M" "M" "M" ...
## $ s10tmin : num NA NA NA NA NA NA NA NA NA NA ...
## $ f.s10tmin: chr "M" "M" "M" "M" ...
Classes in R
The structure results above let us know that the attributes in our data.frame
are stored as several different data types or classes as follows:
- chr - Character: It holds strings that are composed of letters and words. Character class data can not be interpreted numerically - that is to say we can not perform math on these values even if they contain only numbers.
- int - Integer: It holds numbers that are whole integers without decimals. Mathematical operations can be performed on integers.
- num - Numeric: It accepts data that are a wide variety of numeric formats including decimals (floating point values) and integers. Numeric also accept larger numbers than int will.
Storing variables using different classes is a strategic decision by R (and
other programming languages) that optimizes processing and storage. It allows:
- data to be processed more quickly & efficiently.
- the program (R) to minimize the storage size.
Differences Between Classes
Certain functions can be performed on certain data classes and not on others.
For example:
a <- "mouse"
b <- "sparrow"
class(a)
## [1] "character"
class(b)
## [1] "character"
# subtract a-b
a-b
## Error in a - b: non-numeric argument to binary operator
You can not subtract two character values given they are not numbers.
c <- 2
d <- 1
class(c)
## [1] "numeric"
class(d)
## [1] "numeric"
# subtract a-b
c-d
## [1] 1
Additionally, performing summary statistics and other calculations of different types of classes can yield different results.
# create a new object
speciesObserved <- c("speciesb","speciesc","speciesa")
speciesObserved
## [1] "speciesb" "speciesc" "speciesa"
# determine the class
class(speciesObserved)
## [1] "character"
# calculate the minimum
min(speciesObserved)
## [1] "speciesa"
# create numeric object
prec <- c(1,2,5,3,6)
# view class
class(prec)
## [1] "numeric"
# calculate min value
min(prec)
## [1] 1
We can calculate the minimum value for SpeciesObserved, a character
data class, however it does not return a quantitative minimum. It simply
looks for the first element, using alphabetical (rather than numeric) order.
Yet, we can calculate the quantitative minimum value for prec a numeric
data class.
Plot Data Using qplot()
Now that we've got classes down, let's plot one of the metrics in our data,
air temperature -- airt. Given this is a time series dataset, we want to plot
air temperature as it changes over time. We have a date-time column, date, so
let's use that as our x-axis variable and airt as our y-axis variable.
We will use the qplot() (for quick plot) function in the ggplot2 package.
The syntax for qplot() requires the x- and y-axis variables and then the R
object that the variables are stored in.
# quickly plot air temperature
qplot(x=date, y=airt,
data=harMet.daily,
main="Daily Air Temperature\nNEON Harvard Forest Field Site")
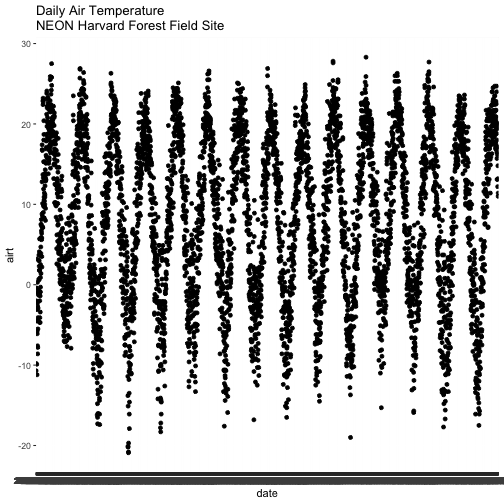
We have successfully plotted some data. However, what is happening on the x-axis?
R is trying to plot EVERY date value in our data, on the x-axis. This makes it hard to read. Why? Let's have a look at the class of the x-axis variable - date.
# View data class for each column that we wish to plot
class(harMet.daily$date)
## [1] "character"
class(harMet.daily$airt)
## [1] "numeric"
In this case, the date column is stored in our data.frame as a character
class. Because it is a character, R does not know how to plot the dates as a
continuous variable. Instead it tries to plot every date value as a text string.
The airt data class is numeric so that metric plots just fine.
Date as a Date-Time Class
We need to convert our date column, which is currently stored as a character
to a date-time class that can be displayed as a continuous variable. Lucky
for us, R has a date class. We can convert the date field to a date class
using as.Date().
# convert column to date class
harMet.daily$date <- as.Date(harMet.daily$date)
# view R class of data
class(harMet.daily$date)
## [1] "Date"
# view results
head(harMet.daily$date)
## [1] "2001-02-11" "2001-02-12" "2001-02-13" "2001-02-14" "2001-02-15"
## [6] "2001-02-16"
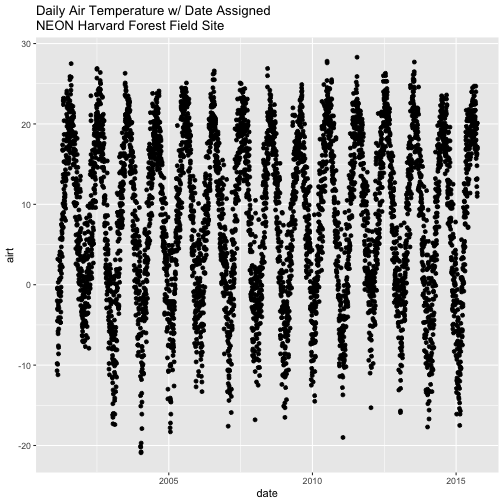
Now that we have adjusted the date, let's plot again. Notice that it plots
much more quickly now that R recognizes date as a date class. R can
aggregate ticks on the x-axis by year instead of trying to plot every day!
# quickly plot the data and include a title using main=""
# In title string we can use '\n' to force the string to break onto a new line
qplot(x=date,y=airt,
data=harMet.daily,
main="Daily Air Temperature w/ Date Assigned\nNEON Harvard Forest Field Site")
- Create a quick plot of the precipitation. Use the full time frame of data available
in the
harMet.dailyobject. - Do precipitation and air temperature have similar annual patterns?
- Create a quick plot examining the relationship between air temperature and precipitation.
Hint: you can modify the X and Y axis labels using xlab="label text" and
ylab="label text".
Get Lesson Code
Time Series 01: Why Metadata Are Important: How to Work with Metadata in Text & EML Format
Authors: Leah A. Wasser, Megan A. Jones, Marisa Guarinello
Last Updated: Apr 8, 2021
This tutorial covers what metadata are, and why we need to work with metadata. It covers the 3 most common metadata formats: text file format, web page format and Ecological Metadata Language (EML).
R Skill Level: Introduction - you've got the basics of R down and understand the general structure of tabular data.
Learning Objectives
After completing this tutorial, you will be able to:
- Import a .csv file and examine the structure of the related R object.
- Use a metadata file to better understand the content of a dataset.
- Explain the importance of including metadata details in your R script.
- Describe what an EML file is.
Things You’ll Need To Complete This Tutorial
You will need the most current version of R and, preferably, RStudio loaded on your computer to complete this tutorial.
Install R Packages
When presented in a workshop, the EML package will be presented as a
demonstration!
If completing EML portion of tutorial on your own, you must
install EML directly from GitHub (the package is in development and is not
yet on CRAN). You will need to install devtools to do this.
-
ggmap:
install.packages("ggmap") -
tmaptools:
install.packages("tmaptools") -
devtools:
install.packages("devtools");library(devtools) -
EML: **must be installed AFTER
devtoolsis loaded in R.install_github("ropensci/EML", build=FALSE, dependencies=c("DEPENDS", "IMPORTS"))
Download Data
NEON Teaching Data Subset: Meteorological Data for Harvard Forest
The data used in this lesson were collected at the National Ecological Observatory Network's Harvard Forest field site. These data are proxy data for what will be available for 30 years on the NEON data portal for the Harvard Forest and other field sites located across the United States.
Set Working Directory: This lesson assumes that you have set your working directory to the location of the downloaded and unzipped data subsets.
An overview of setting the working directory in R can be found here.
R Script & Challenge Code: NEON data lessons often contain challenges that reinforce learned skills. If available, the code for challenge solutions is found in the downloadable R script of the entire lesson, available in the footer of each lesson page.
Understand Our Data
In order to work with any data, we need to understand three things about the data:
- Structure
- Format
- Processing methods
If the data are collected by other people and organizations, we might also need further information about:
- What metrics are included in the dataset
- The units those metrics were stored in
- Explanation of where the metrics are stored in the data and what they are "called" (e.g. what are the column names in a spreadsheet)
- The time range that it covers
- The spatial extent that the data covers
The above information, and more are stored in metadata - data about the data. Metadata is information that describes a dataset and is integral to working with external data (data that we did not collect ourselves).
Metadata Formats
Metadata come in different formats. We will discuss three of those in this tutorial:
-
Ecological Metadata Language (EML): A standardized metadata format stored
in
xmlformat which is machine readable. Metadata has some standards however it's common to encounter metadata stored differently in EML files created by different organizations. - Text file: Sometimes metadata files can be found in text files that are either downloaded with a data product OR that are available separately for the data.
- Directly on a website (HTML / XML): Sometimes data are documented directly in text format, on a web page.
Metadata Stored on a Web Page
The metadata for the data that we are working with for the Harvard Forest field
site are stored in both EML format and on a web page. Let's explore the web
page first.
Let's begin by visiting that page above. At the top of the page, there is a list of data available for Harvard Forest. NOTE: hf001-06: daily (metric) since 2001 (preview) is the data that we used in the previous tutorial.
Scroll down to the Overview section on the website. Take note of the information provided in that section and answer the questions in the Challenge below.
Explore the metadata stored on the Harvard Forest LTER web page. Answer the following questions.
- What is the time span of the data available for this site?
- You have some questions about these data. Who is the lead investigator / who do you contact for more information? And how do you contact them?
- Where is this field site located? How is the site location information stored in the metadata? Is there any information that may be useful to you viewing the location? HINT: What if you were not familiar with Harvard as a site / from another country, etc?
- Field Site Information: What is the elevation for the site? What is the dominant vegetation cover for the site? HINT: Is dominant vegetation easy to find in the metadata?
- How is snow dealt with in the precipitation data?
- Are there some metadata attributes that might be useful to access in a script
in R or
Pythonrather than viewed on a web page? HINT: Can you answer all of the questions above from the information provided on this website? Is there additional information that you might want to find on the page?
View Metadata For Metrics of Interest
For this tutorial series, we are interested in the drivers of plant phenology - specifically air and soil temperature, precipitation and photosynthetically active radiation (PAR). Let's look for descriptions of these variables in the metadata and determine several key attributes that we will need prior to working with the data.
View the metadata at the URL above. Answer the following questions about the Harvard Forest LTER data - hf001-10: 15-minute (metric) since 2005:
- What is the column heading name where each variable (air temperature, soil temperature, precipitation and PAR) is stored?
- What are the units that each variable are stored in?
- What is the frequency of measurement collected for each and how are noData values stored?
- Where is the date information stored (in what field) and what timezone are the dates stored in?
Why Metadata on a Web Page Is Not Ideal
It is nice to have a web page that displays metadata information, however accessing metadata on a web page is difficult:
- If the web page URL changes or the site goes down, the information is lost.
- It's also more challenging to access metadata in text format on a web page programatically - like using R as an interface - which we often want to do when working with larger datasets.
A machine readable metadata file is better - especially when we are working with large data and we want to automate and carefully document workflows. The Ecological Metadata Language (EML) is one machine readable metadata format.
Ecological Metadata Language (EML)
While much of the documentation that we need to work with at the Harvard Forest
field site is available directly on the
Harvard Forest Data Archive page,
the website also offers metadata in EML format.
Introduction to EML
The Ecological Metadata Language (EML) is a data specification developed
specifically
to document ecological data. An EML file is created using a XML based format.
This means that content is embedded within hierarchical tags. For example,
the title of a dataset might be embedded in a <title> tag as follows:
<title>Fisher Meteorological Station at Harvard Forest since 2001</title>
Similarly, the creator of a dataset is also be found in a hierarchical tag structure.
<creator>
<individualName>
<givenName>Emery</givenName>
<surName>Boose</surName>
</individualName>
</creator>
The EML package for R is designed to read and allow users to work with EML
formatted metadata. In this tutorial, we demonstrate how we can use EML in an
automated workflow.
NOTE: The EML package is still being developed, therefore we will not
explicitly teach all details of how to use it. Instead, we will provide
an example of how you can access EML files programmatically and background
information so that you can further explore EML and the EML package if you
need to work with it further.
EML Terminology
Let's first discuss some basic EML terminology. In the context of EML, each EML file documents a dataset. This dataset may consist of one or more files that contain the data in data tables. In the case of our tabular meteorology data, the structure of our EML file includes:
- The dataset. A dataset may contain one or more data tables associated with it that may contain different types of related information. For this Harvard Forest meteorological data, the data tables contain tower measurements including precipitation and temperature, that are aggregated at various time intervals (15 minute, daily, etc) and that date back to 2001.
- The data tables. Data tables refer to the actual data that make up the dataset. For the Harvard Forest data, each data table contains a suite of meteorological metrics, including precipitation and temperature (and associated quality flags), that are aggregated at a particular time interval (e.g. one data table contains monthly average data, another contains 15 minute averaged data, etc).
Work With EML in R
To begin, we will load the EML package directly from ROpenSci's Git repository.
# install R EML tool
# load devtools (if you need to install "EML")
#library("devtools")
# IF YOU HAVE NOT DONE SO ALREADY: install EML from github -- package in
# development; not on CRAN
#install_github("ropensci/EML", build=FALSE, dependencies=c("DEPENDS", "IMPORTS"))
# load ROpenSci EML package
library(EML)
# load ggmap for mapping
library(ggmap)
# load tmaptools for mapping
library(tmaptools)
Next, we will read in the LTER EML file - directly from the online URL using
eml_read. This file documents multiple data products that can be downloaded.
Check out the
Harvard Forest Data Archive Page for Fisher Meteorological Station
for more on this dataset and to download the archive files directly.
Note that because this EML file is large, it takes a few seconds for the file to load.
# data location
# http://harvardforest.fas.harvard.edu:8080/exist/apps/datasets/showData.html?id=hf001
# table 4 http://harvardforest.fas.harvard.edu/data/p00/hf001/hf001-04-monthly-m.csv
# import EML from Harvard Forest Met Data
# note, for this particular tutorial, we will work with an abridged version of the file
# that you can access directly on the harvard forest website. (see comment below)
eml_HARV <- read_eml("https://harvardforest1.fas.harvard.edu/sites/harvardforest.fas.harvard.edu/files/data/eml/hf001.xml")
# import a truncated version of the eml file for quicker demonstration
# eml_HARV <- read_eml("http://neonscience.github.io/NEON-R-Tabular-Time-Series/hf001-revised.xml")
# view size of object
object.size(eml_HARV)
## 1299568 bytes
# view the object class
class(eml_HARV)
## [1] "emld" "list"
The eml_read function creates an EML class object. This object can be
accessed using slots in R ($) rather than a typical subset [] approach.
Explore Metadata Attributes
We can begin to explore the contents of our EML file and associated data that it
describes. Let's start at the dataset level. We can use slots to view
the contact information for the dataset and a description of the methods.
# view the contact name listed in the file
eml_HARV$dataset$creator
## $individualName
## $individualName$givenName
## [1] "Emery"
##
## $individualName$surName
## [1] "Boose"
# view information about the methods used to collect the data as described in EML
eml_HARV$dataset$methods
## $methodStep
## $methodStep$description
## $methodStep$description$section
## $methodStep$description$section[[1]]
## [1] "<title>Observation periods</title><para>15-minute: 15 minutes, ending with given time. Hourly: 1 hour, ending with given time. Daily: 1 day, midnight to midnight. All times are Eastern Standard Time.</para>"
##
## $methodStep$description$section[[2]]
## [1] "<title>Instruments</title><para>Air temperature and relative humidity: Vaisala HMP45C (2.2m above ground). Precipitation: Met One 385 heated rain gage (top of gage 1.6m above ground). Global solar radiation: Licor LI200X pyranometer (2.7m above ground). PAR radiation: Licor LI190SB quantum sensor (2.7m above ground). Net radiation: Kipp and Zonen NR-LITE net radiometer (5.0m above ground). Barometric pressure: Vaisala CS105 barometer. Wind speed and direction: R.M. Young 05103 wind monitor (10m above ground). Soil temperature: Campbell 107 temperature probe (10cm below ground). Data logger: Campbell Scientific CR10X.</para>"
##
## $methodStep$description$section[[3]]
## [1] "<title>Instrument and flag notes</title><para>Air temperature. Daily air temperature is estimated from other stations as needed to complete record.</para><para>Precipitation. Daily precipitation is estimated from other stations as needed to complete record. Delayed melting of snow and ice (caused by problems with rain gage heater or heavy precipitation) is noted in log - daily values are corrected if necessary but 15-minute values are not. The gage may underestimate actual precipitation under windy or cold conditions.</para><para>Radiation. Whenever possible, snow and ice are removed from radiation instruments after precipitation events. Depth of snow or ice on instruments and time of removal are noted in log, but values are not corrected or flagged.</para><para>Wind speed and direction. During ice storms, values are flagged as questionable when there is evidence (from direct observation or the 15-minute record) that ice accumulation may have affected the instrument's operation.</para>"
Identify & Map Data Location
Looking at the coverage for our data, there is only one unique x and y value.
This suggests that our data were collected at (x,y) one point location. We know
this is a tower so a point location makes sense. Let's grab the x and y
coordinates and create a quick context map. We will use ggmap to create our
map.
NOTE: If this were a rectangular extent, we'd want the bounding box not just the point. This is important if the data in raster, HDF5, or a similar format. We need the extent to properly geolocate and process the data.
# grab x coordinate from the coverage information
XCoord <- eml_HARV$dataset$coverage$geographicCoverage$boundingCoordinates$westBoundingCoordinate[[1]]
# grab y coordinate from the coverage information
YCoord <- eml_HARV$dataset$coverage$geographicCoverage$boundingCoordinates$northBoundingCoordinate[[1]]
# make a map and add the xy coordinates to it
ggmap(get_stamenmap(rbind(as.numeric(paste(geocode_OSM("Massachusetts")$bbox))), zoom = 11, maptype=c("toner")), extent=TRUE) + geom_point(aes(x=as.numeric(XCoord),y=as.numeric(YCoord)),
color="darkred", size=6, pch=18)
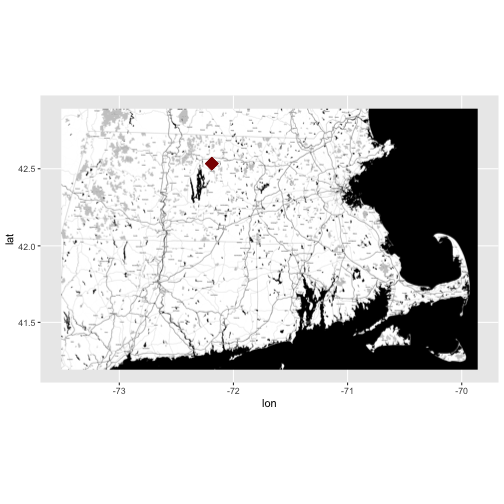
- Learn more about ggmap: A nice cheatsheet created by NCEAS
The above example, demonstrated how we can extract information from an EML
document and use it programatically in R! This is just the beginning of what
we can do!
Metadata For Your Own Data
Now, imagine that you are working with someone else's data and you don't have a metadata file associated with it? How do you know what units the data were in? How the data were collected? The location that the data covers? It is equally important to create metadata for your own data, to make your data more efficiently "shareable".
Get Lesson Code
Time Series 02: Dealing With Dates & Times in R - as.Date, POSIXct, POSIXlt
Authors: Megan A. Jones, Marisa Guarinello, Courtney Soderberg, Leah A. Wasser
Last Updated: May 13, 2021
This tutorial explores working with date and time field in R. We will overview
the differences between as.Date, POSIXct and POSIXlt as used to convert
a date / time field in character (string) format to a date-time format that is
recognized by R. This conversion supports efficient plotting, subsetting and
analysis of time series data.
Learning Objectives
After completing this tutorial, you will be able to:
- Describe various date-time classes and data structure in R.
- Explain the difference between
POSIXctandPOSIXltdata classes are and why POSIXct may be preferred for some tasks. - Convert a column containing date-time information in character format to a date-time R class.
- Convert a date-time column to different date-time classes.
- Write out a date-time class object in different ways (month-day, month-day-year, etc).
Things You’ll Need To Complete This Tutorials
You will need the most current version of R and, preferably, RStudio loaded on your computer to complete this tutorial.
Install R Packages
-
lubridate:
install.packages("lubridate")
More on Packages in R – Adapted from Software Carpentry.
Download Data
NEON Teaching Data Subset: Meteorological Data for Harvard Forest
The data used in this lesson were collected at the National Ecological Observatory Network's Harvard Forest field site. These data are proxy data for what will be available for 30 years on the NEON data portal for the Harvard Forest and other field sites located across the United States.
Set Working Directory: This lesson assumes that you have set your working directory to the location of the downloaded and unzipped data subsets.
An overview of setting the working directory in R can be found here.
R Script & Challenge Code: NEON data lessons often contain challenges that reinforce learned skills. If available, the code for challenge solutions is found in the downloadable R script of the entire lesson, available in the footer of each lesson page.
The Data Approach
Intro to Time Series Data in R tutorial
we imported a time series dataset in .csv format into R. We learned how to
quickly plot these data by converting the date column to an R Date class.
In this tutorial we will explore how to work with a column that contains both a
date AND a time stamp.
We will use functions from both base R and the lubridate package to work
with date-time data classes.
# Load packages required for entire script
library(lubridate) #work with dates
#Set the working directory and place your downloaded data there
wd <- "~/Git/data/"
Import CSV File
First, let's import our time series data. We are interested in temperature,
precipitation and photosynthetically active radiation (PAR) - metrics that are
strongly associated with vegetation green-up and brown down (phenology or
phenophase timing). We will use the hf001-10-15min-m.csv file
that contains atmospheric data for the NEON Harvard Forest field site,
aggregated at 15-minute intervals. Download the dataset for these exercises here.
# Load csv file of 15 min meteorological data from Harvard Forest
# contained within the downloaded directory, or available for download
# directly from:
# https://harvardforest.fas.harvard.edu/data/p00/hf001/hf001-10-15min-m.csv
# Factors=FALSE so strings, series of letters/words/numerals, remain characters
harMet_15Min <- read.csv(
file=paste0(wd,"NEON-DS-Met-Time-Series/HARV/FisherTower-Met/hf001-10-15min-m.csv"),
stringsAsFactors = FALSE)
Date and Time Data
Let's revisit the data structure of our harMet_15Min object. What is the class
of the date-time column?
# view column data class
class(harMet_15Min$datetime)
## [1] "character"
# view sample data
head(harMet_15Min$datetime)
## [1] "2005-01-01T00:15" "2005-01-01T00:30" "2005-01-01T00:45"
## [4] "2005-01-01T01:00" "2005-01-01T01:15" "2005-01-01T01:30"
Our datetime column is stored as a character class. We need to convert it to
date-time class. What happens when we use the as.Date method that we learned
about in the
Intro to Time Series Data in R tutorial?
# convert column to date class
dateOnly_HARV <- as.Date(harMet_15Min$datetime)
# view data
head(dateOnly_HARV)
## [1] "2005-01-01" "2005-01-01" "2005-01-01" "2005-01-01" "2005-01-01"
## [6] "2005-01-01"
When we use as.Date, we lose the time stamp.
Explore Date and Time Classes
R - Date Class - as.Date
As we just saw, the as.Date format doesn't store any time information. When we
use theas.Date method to convert a date stored as a character class to an R
date class, it will ignore all values after the date string.
# Convert character data to date (no time)
myDate <- as.Date("2015-10-19 10:15")
str(myDate)
## Date[1:1], format: "2015-10-19"
# what happens if the date has text at the end?
myDate2 <- as.Date("2015-10-19Hello")
str(myDate2)
## Date[1:1], format: "2015-10-19"
As we can see above the as.Date() function will convert the characters that it
recognizes to be part of a date into a date class and ignore all other
characters in the string.
R - Date-Time - The POSIX classes
If we have a column containing both date and time we need to use a class that
stores both date AND time. Base R offers two closely related classes for date
and time: POSIXct and POSIXlt.
POSIXct
The as.POSIXct method converts a date-time string into a POSIXct class.
# Convert character data to date and time.
timeDate <- as.POSIXct("2015-10-19 10:15")
str(timeDate)
## POSIXct[1:1], format: "2015-10-19 10:15:00"
timeDate
## [1] "2015-10-19 10:15:00 MDT"
as.POSIXct stores both a date and time with an associated time zone. The
default time zone selected, is the time zone that your computer is set to which
is most often your local time zone.
POSIXct stores date and time in seconds with the number of seconds beginning
at 1 January 1970. Negative numbers are used to store dates prior to 1970.
Thus, the POSIXct format stores each date and time a single value in units of
seconds. Storing the data this way, optimizes use in data.frames and speeds up
computation, processing and conversion to other formats.
# to see the data in this 'raw' format, i.e., not formatted according to the
# class type to show us a date we recognize, use the `unclass()` function.
unclass(timeDate)
## [1] 1445271300
## attr(,"tzone")
## [1] ""
Here we see the number of seconds from 1 January 1970 to 9 October 2015
(1445271300). Also, we see that a time zone (tzone) is associated with the value in seconds.
POSIXlt
The POSIXct format is optimized for storage and computation. However, humans
aren't quite as computationally efficient as computers! Also, we often want to
quickly extract some portion of the data (e.g., months). The POSIXlt class
stores date and time information in a format that we are used to seeing (e.g.,
second, min, hour, day of month, month, year, numeric day of year, etc).
# Convert character data to POSIXlt date and time
timeDatelt<- as.POSIXlt("2015-10-19 10:15")
str(timeDatelt)
## POSIXlt[1:1], format: "2015-10-19 10:15:00"
timeDatelt
## [1] "2015-10-19 10:15:00 MDT"
unclass(timeDatelt)
## $sec
## [1] 0
##
## $min
## [1] 15
##
## $hour
## [1] 10
##
## $mday
## [1] 19
##
## $mon
## [1] 9
##
## $year
## [1] 115
##
## $wday
## [1] 1
##
## $yday
## [1] 291
##
## $isdst
## [1] 1
##
## $zone
## [1] "MDT"
##
## $gmtoff
## [1] NA
When we convert a string to POSIXlt, and view it in R, it still looks
similar to the POSIXct format. However, unclass() shows us that the data are
stored differently. The POSIXlt class stores the hour, minute, second, day,
month, and year separately.
Months in POSIXlt
POSIXlt has a few quirks. First, the month values stored in the POSIXlt
object use zero-based indexing. This means that month #1 (January) is stored
as 0, and month #2 (February) is stored as 1. Notice in the output above,
October is stored as the 9th month ($mon = 9).
Years in POSIXlt
Years are also stored differently in the POSIXlt class. Year values are stored
using a base index value of 1900. Thus, 2015 is stored as 115 ($year = 115
- 115 years since 1900).
Having dates classified as separate components makes POSIXlt computationally
more resource intensive to use in data.frames. This slows things down! We will
thus use POSIXct for this tutorial.
Convert to Date-time Class
When we convert from a character to a date-time class we need to tell R how
the date and time information are stored in each string. To do this, we can use
format=. Let's have a look at one of our date-time strings to determine it's
format.
# view one date-time field
harMet_15Min$datetime[1]
## [1] "2005-01-01T00:15"
Looking at the results above, we see that our data are stored in the format:
Year-Month-Day "T" Hour:Minute (2005-01-01T00:15). We can use this information
to populate our format string using the following designations for the
components of the date-time data:
- %Y - year
- %m - month
- %d - day
- %H:%M - hours:minutes
The "T" inserted into the middle of datetime isn't a standard character for
date and time, however we can tell R where the character is so it can ignore
it and interpret the correct date and time as follows:
format="%Y-%m-%dT%H:%M".
# convert single instance of date/time in format year-month-day hour:min:sec
as.POSIXct(harMet_15Min$datetime[1],format="%Y-%m-%dT%H:%M")
## [1] "2005-01-01 00:15:00 MST"
## The format of date-time MUST match the specified format or the data will not
# convert; see what happens when you try it a different way or without the "T"
# specified
as.POSIXct(harMet_15Min$datetime[1],format="%d-%m-%Y%H:%M")
## [1] NA
as.POSIXct(harMet_15Min$datetime[1],format="%Y-%m-%d%H:%M")
## [1] NA
Using the syntax we've learned, we can convert the entire datetime column into
POSIXct class.
new.date.time <- as.POSIXct(harMet_15Min$datetime,
format="%Y-%m-%dT%H:%M" #format time
)
# view output
head(new.date.time)
## [1] "2005-01-01 00:15:00 MST" "2005-01-01 00:30:00 MST"
## [3] "2005-01-01 00:45:00 MST" "2005-01-01 01:00:00 MST"
## [5] "2005-01-01 01:15:00 MST" "2005-01-01 01:30:00 MST"
# what class is the output
class(new.date.time)
## [1] "POSIXct" "POSIXt"
About Time Zones
Above, we successfully converted our data into a date-time class. However, what
timezone is the output new.date.time object that we created using?
2005-04-15 03:30:00 MDT
It appears as if our data were assigned MDT (which is the timezone of the computer where these tutorials were processed originally - in Colorado - Mountain Time). However, we know from the metadata, explored in the Why Metadata Are Important: How to Work with Metadata in Text & EML Format tutorial, that these data were stored in Eastern Standard Time.
Assign Time Zone
When we convert a date-time formatted column to POSIXct format, we need to
assign an associated time zone. If we don't assign a time zone,R will
default to the local time zone that is defined on your computer.
There are individual designations for different time zones and time zone
variants (e.g., does the time occur during daylight savings time).
The code for the Eastern time zone that is the closest match to the NEON Harvard
Forest field site is America/New_York. Let's convert our datetime field
one more time, and define the associated timezone (tz=).
# assign time zone to just the first entry
as.POSIXct(harMet_15Min$datetime[1],
format = "%Y-%m-%dT%H:%M",
tz = "America/New_York")
## [1] "2005-01-01 00:15:00 EST"
The output above, shows us that the time zone is now correctly set as EST.
Convert to Date-time Data Class
Now, using the syntax that we learned above, we can convert the entire
datetime column to a POSIXct class.
# convert to POSIXct date-time class
harMet_15Min$datetime <- as.POSIXct(harMet_15Min$datetime,
format = "%Y-%m-%dT%H:%M",
tz = "America/New_York")
# view structure and time zone of the newly defined datetime column
str(harMet_15Min$datetime)
## POSIXct[1:376800], format: "2005-01-01 00:15:00" "2005-01-01 00:30:00" ...
tz(harMet_15Min$datetime)
## [1] "America/New_York"
Now that our datetime data are properly identified as a POSIXct date-time
data class we can continue on and look at the patterns of precipitation,
temperature, and PAR through time.
Get Lesson Code
Time Series 03: Cleaning & Subsetting Time Series Data in R - NoData Values & Subset by Date
Authors: Megan A. Jones, Marisa Guarinello, Courtney Soderberg, Leah A. Wasser
Last Updated: May 13, 2021
This tutorial explores how to deal with NoData values encountered in a time
series dataset, in R. It also covers how to subset large files by date and
export the results to a .csv (text) file.
Learning Objectives
After completing this tutorial, you will be able to:
- Subset data by date.
- Search for NA or missing data values.
- Describe different possibilities on how to deal with missing data.
Things You’ll Need To Complete This Tutorial
You will need the most current version of R and, preferably, RStudio loaded on your computer to complete this tutorial.
Install R Packages
-
lubridate:
install.packages("lubridate") -
ggplot2:
install.packages("ggplot2")
More on Packages in R – Adapted from Software Carpentry.
Download Data
NEON Teaching Data Subset: Meteorological Data for Harvard Forest
The data used in this lesson were collected at the National Ecological Observatory Network's Harvard Forest field site. These data are proxy data for what will be available for 30 years on the NEON data portal for the Harvard Forest and other field sites located across the United States.
Set Working Directory: This lesson assumes that you have set your working directory to the location of the downloaded and unzipped data subsets.
An overview of setting the working directory in R can be found here.
R Script & Challenge Code: NEON data lessons often contain challenges that reinforce learned skills. If available, the code for challenge solutions is found in the downloadable R script of the entire lesson, available in the footer of each lesson page.
Cleaning Time Series Data
It is common to encounter, large files containing more
data than we need for our analysis. It is also common to encounter NoData
values that we need to account for when analyzing our data.
In this tutorial, we'll learn how to both manage NoData values and also
subset and export a portion of an R object as a new .csv file.
In this tutorial, we will work with atmospheric data, containing air temperature, precipitation, and photosynthetically active radiation (PAR) data - metrics that are key drivers of phenology. Our study area is the NEON Harvard Forest Field Site.
Import Timeseries Data
We will use the lubridate and ggplot2 packages. Let's load those first.
If you have not already done so, import the hf001-10-15min-m.csv file, which
contains atmospheric data for Harvard Forest. Convert the datetime column
to a POSIXct class as covered in the tutorial:
Dealing With Dates & Times in R - as.Date, POSIXct, POSIXlt.
# Load packages required for entire script
library(lubridate) # work with dates
library(ggplot2) # plotting
# set working directory to ensure R can find the file we wish to import
wd <- "~/Git/data/"
# Load csv file containing 15 minute averaged atmospheric data
# for the NEON Harvard Forest Field Site
# Factors=FALSE so data are imported as numbers and characters
harMet_15Min <- read.csv(
file=paste0(wd,"NEON-DS-Met-Time-Series/HARV/FisherTower-Met/hf001-10-15min-m.csv"),
stringsAsFactors = FALSE)
# convert to POSIX date time class - US Eastern Time Zone
harMet_15Min$datetime <- as.POSIXct(harMet_15Min$datetime,
format = "%Y-%m-%dT%H:%M",
tz = "America/New_York")
Subset by Date
Our .csv file contains nearly a decade's worth of data which makes for a large
file. The time period we are interested in for our study is:
- Start Time: 1 January 2009
- End Time: 31 Dec 2011
Let's subset the data to only contain these three years. We can use the
subset() function, with the syntax:
NewObject <- subset ( ObjectToBeSubset, CriteriaForSubsetting ) .
We will set our criteria to be any datetime that:
- Is greater than or equal to 1 Jan 2009 at 0:00 AND
- Is less than or equal to 31 Dec 2011 at 23:59.
We also need to specify the timezone so R can handle daylight savings and
leap year.
# subset data - 2009-2011
harMet15.09.11 <- subset(harMet_15Min,
datetime >= as.POSIXct('2009-01-01 00:00',
tz = "America/New_York") &
datetime <= as.POSIXct('2011-12-31 23:59',
tz = "America/New_York"))
# View first and last records in the object
head(harMet15.09.11[1])
## datetime
## 140255 2009-01-01 00:00:00
## 140256 2009-01-01 00:15:00
## 140257 2009-01-01 00:30:00
## 140258 2009-01-01 00:45:00
## 140259 2009-01-01 01:00:00
## 140260 2009-01-01 01:15:00
tail(harMet15.09.11[1])
## datetime
## 245369 2011-12-31 22:30:00
## 245370 2011-12-31 22:45:00
## 245371 2011-12-31 23:00:00
## 245372 2011-12-31 23:15:00
## 245373 2011-12-31 23:30:00
## 245374 2011-12-31 23:45:00
It worked! The first entry is 1 January 2009 at 00:00 and the last entry is 31 December 2011 at 23:45.
Export data.frame to .CSV
We can export this subset in .csv format to use in other analyses or to
share with colleagues using write.csv.
# write harMet15 subset data to .csv
write.csv(harMet15.09.11,
file=paste0(wd,"Met_HARV_15min_2009_2011.csv"))
-
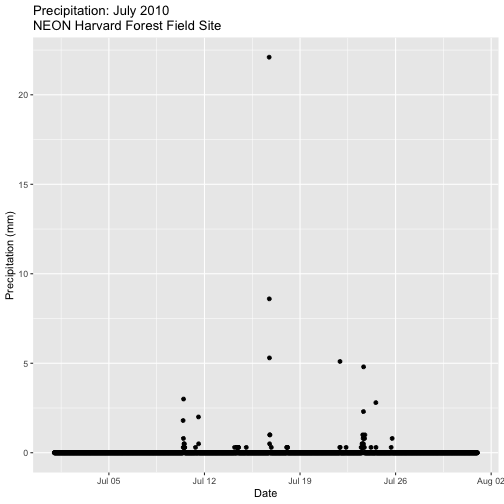
Create a plot of precipitation for the month of July 2010 in Harvard Forest. Be sure to label x and y axes. Also be sure to give your plot a title.
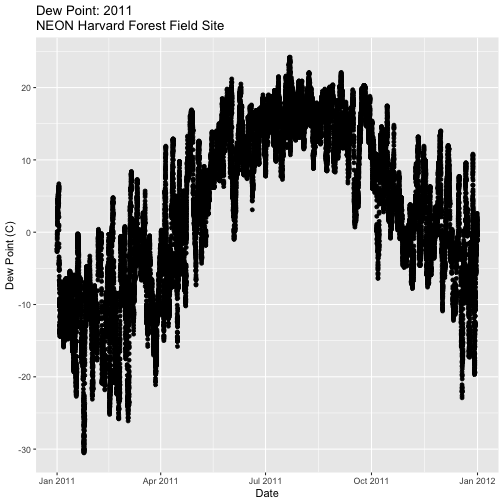
-
Create a plot of dew point (
dewp) for the year 2011 at Harvard Forest.
Bonus challenge: Complete this challenge using the available daily data instead of the 15-minute data. What will need to change in your subsetting code?
Managing Missing Data: NoData values
Find NoData Values
If we are lucky when working with external data, the NoData value is clearly
specified
in the metadata. No data values can be stored differently:
-
NA / NaN: Sometimes this value is
NAorNaN(not a number). -
A Designated Numeric Value (e.g. -9999): Character strings such as
NAcan not always be stored along side of numeric values in some file formats. Sometimes you'll encounter numeric placeholders fornoDatavalues such as-9999(a value often used in the GIS / Remote Sensing and Micrometeorology domains. -
Blank Values: sometimes
noDatavalues are left blank. Blanks are particularly problematic because we can't be certain if a data value is purposefully missing (not measured that day or a bad measurement) or if someone unintentionally deleted it.
Because the actual value used to designate missing data can vary depending upon
what data we are working with, it is important to always check the metadata for
the files associated NoData value. If the value is NA, we are in luck, R
will recognize and flag this value as NoData. If the value is numeric (e.g.,
-9999), then we might need to assign this value to NA.
In the
Why Metadata Are Important: How to Work with Metadata in Text & EML Format tutorial,
we viewed the metadata for these data and discovered that missing values are
designated using NA - a common NoData value placeholder.
Excerpt from the metadata:
airt: average air temperature. Average of daily averages. (unit: celsius / missing value: NA)
Check For NoData Values
We can quickly check for NoData values in our data using theis.na()
function. By asking for the sum() of is.na() we can see how many NA/ missing
values we have.
# Check for NA values
sum(is.na(harMet15.09.11$datetime))
## [1] 0
sum(is.na(harMet15.09.11$airt))
## [1] 2
# view rows where the air temperature is NA
harMet15.09.11[is.na(harMet15.09.11$airt),]
## datetime jd airt f.airt rh f.rh dewp f.dewp prec
## 158360 2009-07-08 14:15:00 189 NA M NA M NA M 0
## 203173 2010-10-18 09:30:00 291 NA M NA M NA M 0
## f.prec slrr f.slrr parr f.parr netr f.netr bar f.bar wspd
## 158360 290 485 139 NA M 2.1
## 203173 NA M NA M NA M NA M NA
## f.wspd wres f.wres wdir f.wdir wdev f.wdev gspd f.gspd s10t
## 158360 1.8 86 29 5.2 20.7
## 203173 M NA M NA M NA M NA M 10.9
## f.s10t
## 158360
## 203173
The results above tell us there are NoData values in the airt column.
However, there are NoData values in other variables.
How many NoData values are in the precipitation (prec) and PAR (parr)
columns of our data?
Deal with NoData Values
When we encounter NoData values (blank, NaN, -9999, etc.) in our data we
need to decide how to deal with them. By default R treats NoData values
designated with a NA as a missing value rather than a zero. This is good, as a
value of zero (no rain today) is not the same as missing data (e.g. we didn't
measure the amount of rainfall today).
How we deal with NoData values will depend on:
- the data type we are working with
- the analysis we are conducting
- the significance of the gap or missing value
Many functions in R contains a na.rm= option which will allow you to tell R
to ignore NA values in your data when performing calculations.
To Gap Fill? Or Not?
Sometimes we might need to "gap fill" our data. This means we will interpolate or estimate missing values often using statistical methods. Gap filling can be complex and is beyond the scope of this tutorial. The take away from this is simply that it is important to acknowledge missing values in your data and to carefully consider how you wish to account for them during analysis.
Other resources:
- R code for dealing with missing data: Quick-R: Missing Data
Managing NoData Values in Our Data
For this tutorial, we are exploring the patterns of precipitation,
and temperature as they relate to green-up and brown-down of vegetation at
Harvard Forest. To view overall trends during these early exploration stages, it
is okay for us to leave out the NoData values in our plots.
NoData Values Can Impact Calculations
It is important to consider NoData values when performing calculations on our
data. For example, R will not properly calculate certain functions if there
are NA values in the data, unless we explicitly tell it to ignore them.
# calculate mean of air temperature
mean(harMet15.09.11$airt)
## [1] NA
# are there NA values in our data?
sum(is.na(harMet15.09.11$airt))
## [1] 2
R will not return a value for the mean as there NA values in the air
temperature column. Because there are only 2 missing values (out of 105,108) for
air temperature, we aren't that worried about a skewed 3 year mean. We can tell
R to ignore noData values in the mean calculations using na.rm=
(NA.remove).
# calculate mean of air temperature, ignore NA values
mean(harMet15.09.11$airt,
na.rm=TRUE)
## [1] 8.467904
We now see that the 3-year average air temperature is 8.5°C.
- Import the Daily Meteorological data from the Harvard Forest (if you haven't already done so in the Intro to Time Series Data in R tutorial.)
- Check the metadata to see what the column names are for the variable of interest (precipitation, air temperature, PAR, day and time ).
- If needed, convert the data class of different columns.
- Check for missing data and decide what to do with any that exist.
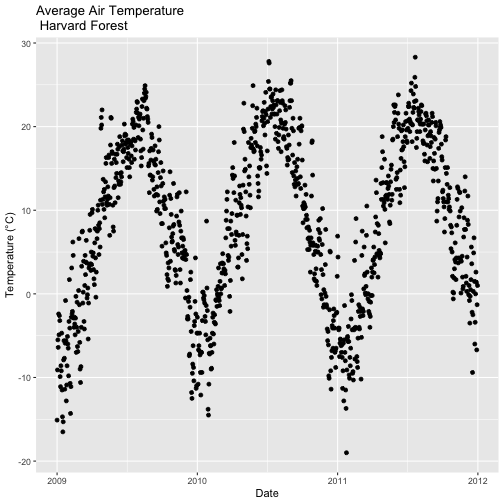
- Subset out the data for the duration of our study: 2009-2011. Name the object "harMetDaily.09.11".
- Export the subset to a
.csvfile. - Create a plot of Daily Air Temperature for 2009-2011. Be sure to label, x- and y-axes. Also give the plot a title!
Get Lesson Code
Time Series 04: Subset and Manipulate Time Series Data with dplyr
Authors: Megan A. Jones, Marisa Guarinello, Courtney Soderberg, Leah A. Wasser
Last Updated: May 13, 2021
In this tutorial, we will use the group_by, summarize and mutate functions
in the dplyr package to efficiently manipulate atmospheric data collected at
the NEON Harvard Forest Field Site. We will use pipes to efficiently perform
multiple tasks within a single chunk of code.
Learning Objectives
After completing this tutorial, you will be able to:
- Explain several ways to manipulate data using functions in the
dplyrpackage in R. - Use
group-by(),summarize(), andmutate()functions. - Write and understand R code with pipes for cleaner, efficient coding.
- Use the
year()function from thelubridatepackage to extract year from a date-time class variable.
Things You’ll Need To Complete This Tutorial
You will need the most current version of R and, preferably, RStudio loaded on your computer to complete this tutorial.
Install R Packages
-
lubridate:
install.packages("lubridate") -
dplyr:
install.packages("dplyr") -
ggplot2:
install.packages("ggplot2")
More on Packages in R – Adapted from Software Carpentry.
Download Data
NEON Teaching Data Subset: Meteorological Data for Harvard Forest
The data used in this lesson were collected at the National Ecological Observatory Network's Harvard Forest field site. These data are proxy data for what will be available for 30 years on the NEON data portal for the Harvard Forest and other field sites located across the United States.
Set Working Directory: This lesson assumes that you have set your working directory to the location of the downloaded and unzipped data subsets.
An overview of setting the working directory in R can be found here.
R Script & Challenge Code: NEON data lessons often contain challenges that reinforce learned skills. If available, the code for challenge solutions is found in the downloadable R script of the entire lesson, available in the footer of each lesson page.
Additional Resources
- NEON Data Skills tutorial on spatial data and piping with dplyr
- Data Carpentry lesson's on Aggregating and Analyzing Data with dplyr
-
dplyrpackage description. -
RStudio Introduction to
dplyr
Introduction to dplyr
The dplyr package simplifies and increases efficiency of complicated yet
commonly performed data "wrangling" (manipulation / processing) tasks. It uses
the data_frame object as both an input and an output.
Load the Data
We will need the lubridate and the dplyr packages to complete this tutorial.
We will also use the 15-minute average atmospheric data subsetted to 2009-2011 for the NEON Harvard Forest Field Site. This subset was created in the Subsetting Time Series Data tutorial.
If this object isn't already created, please load the .csv version:
Met_HARV_15min_2009_2011.csv. Be
sure to convert the date-time column to a POSIXct class after the .csv is
loaded.
# it's good coding practice to load packages at the top of a script
library(lubridate) # work with dates
library(dplyr) # data manipulation (filter, summarize, mutate)
library(ggplot2) # graphics
library(gridExtra) # tile several plots next to each other
library(scales)
# set working directory to ensure R can find the file we wish to import
wd <- "~/Git/data/"
# 15-min Harvard Forest met data, 2009-2011
harMet15.09.11<- read.csv(
file=paste0(wd,"NEON-DS-Met-Time-Series/HARV/FisherTower-Met/Met_HARV_15min_2009_2011.csv"),
stringsAsFactors = FALSE)
# convert datetime to POSIXct
harMet15.09.11$datetime<-as.POSIXct(harMet15.09.11$datetime,
format = "%Y-%m-%d %H:%M",
tz = "America/New_York")
Explore The Data
Remember that we are interested in the drivers of phenology including - air temperature, precipitation, and PAR (photosynthetic active radiation - or the amount of visible light). Using the 15-minute averaged data, we could easily plot each of these variables.
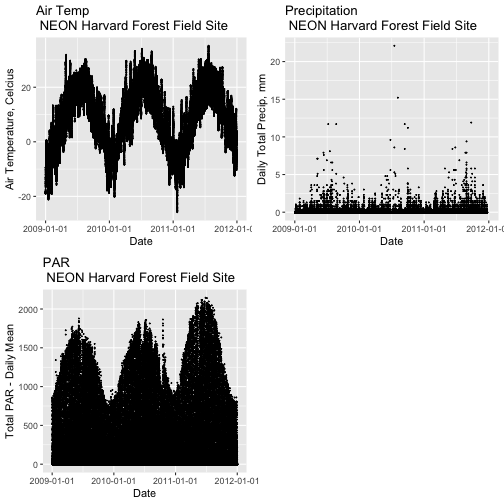
However, summarizing the data at a coarser scale (e.g., daily, weekly, by season, or by year) may be easier to visually interpret during initial stages of data exploration.
Extract Year from a Date-Time Column
To summarize by year efficiently, it is helpful to have a year column that we
can use to group by. We can use the lubridate function year() to extract
the year only from a date-time class R column.
# create a year column
harMet15.09.11$year <- year(harMet15.09.11$datetime)
Using names() we see that we now have a year column in our data_frame.
# check to make sure it worked
names(harMet15.09.11)
## [1] "X" "datetime" "jd" "airt" "f.airt" "rh"
## [7] "f.rh" "dewp" "f.dewp" "prec" "f.prec" "slrr"
## [13] "f.slrr" "parr" "f.parr" "netr" "f.netr" "bar"
## [19] "f.bar" "wspd" "f.wspd" "wres" "f.wres" "wdir"
## [25] "f.wdir" "wdev" "f.wdev" "gspd" "f.gspd" "s10t"
## [31] "f.s10t" "year"
str(harMet15.09.11$year)
## num [1:105108] 2009 2009 2009 2009 2009 ...
Now that we have added a year column to our data_frame, we can use dplyr to
summarize our data.
Manipulate Data using dplyr
Let's start by extracting a yearly air temperature value for the Harvard Forest data. To calculate a yearly average, we need to:
- Group our data by year.
- Calculate the mean precipitation value for each group (ie for each year).
We will use dplyr functions group_by and summarize to perform these steps.
# Create a group_by object using the year column
HARV.grp.year <- group_by(harMet15.09.11, # data_frame object
year) # column name to group by
# view class of the grouped object
class(HARV.grp.year)
## [1] "grouped_df" "tbl_df" "tbl" "data.frame"
The group_by function creates a "grouped" object. We can then use this
grouped object to quickly calculate summary statistics by group - in this case,
year. For example, we can count how many measurements were made each year using
the tally() function. We can then use the summarize function to calculate
the mean air temperature value each year. Note that "summarize" is a common
function name across many different packages. If you happen to have two packages
loaded at the same time that both have a "summarize" function, you may not get
the results that you expect. Therefore, we will "disambiguate" our function by
specifying which package it comes from using the double colon notation
dplyr::summarize().
# how many measurements were made each year?
tally(HARV.grp.year)
## # A tibble: 3 x 2
## year n
## <dbl> <int>
## 1 2009 35036
## 2 2010 35036
## 3 2011 35036
# what is the mean airt value per year?
dplyr::summarize(HARV.grp.year,
mean(airt) # calculate the annual mean of airt
)
## # A tibble: 3 x 2
## year `mean(airt)`
## <dbl> <dbl>
## 1 2009 NA
## 2 2010 NA
## 3 2011 8.75
Why did this return a NA value for years 2009 and 2010? We know there are some
values for both years. Let's check our data for NoData values.
# are there NoData values?
sum(is.na(HARV.grp.year$airt))
## [1] 2
# where are the no data values
# just view the first 6 columns of data
HARV.grp.year[is.na(HARV.grp.year$airt),1:6]
## # A tibble: 2 x 6
## X datetime jd airt f.airt rh
## <int> <dttm> <int> <dbl> <chr> <int>
## 1 158360 2009-07-08 14:15:00 189 NA M NA
## 2 203173 2010-10-18 09:30:00 291 NA M NA
It appears as if there are two NoData values (in 2009 and 2010) that are
causing R to return a NA for the mean for those years. As we learned
previously, we can use na.rm to tell R to ignore those values and calculate
the final mean value.
# calculate mean but remove NA values
dplyr::summarize(HARV.grp.year,
mean(airt, na.rm = TRUE)
)
## # A tibble: 3 x 2
## year `mean(airt, na.rm = TRUE)`
## <dbl> <dbl>
## 1 2009 7.63
## 2 2010 9.03
## 3 2011 8.75
Great! We've now used the group_by function to create groups for each year
of our data. We then calculated a summary mean value per year using summarize.
dplyr Pipes
The above steps utilized several steps of R code and created 1 R object -
HARV.grp.year. We can combine these steps using pipes in the dplyr
package.
We can use pipes to string functions or processing steps together. The output
of each step is fed directly into the next step using the syntax: %>%. This
means we don't need to save the output of each intermediate step as a new R
object.
A few notes about piping syntax:
- A pipe begins with the object name that we will be manipulating, in our case
harMet15.09.11. - It then links that object with first manipulation step using
%>%. - Finally, the first function is called, in our case
group_by(year). Note that because we specified the object name in step one above, we can just use the column name
So, we have: harMet15.09.11 %>% group_by(year)
- We can then add an additional function (or more functions!) to our pipe. For
example, we can tell R to
tallyor count the number of measurements per year.
harMet15.09.11 %>% group_by(year) %>% tally()
Let's try it!
# how many measurements were made a year?
harMet15.09.11 %>%
group_by(year) %>% # group by year
tally() # count measurements per year
## # A tibble: 3 x 2
## year n
## <dbl> <int>
## 1 2009 35036
## 2 2010 35036
## 3 2011 35036
Piping allows us to efficiently perform operations on our data_frame in that:
- It allows us to condense our code, without naming intermediate steps.
- The dplyr package is optimized to ensure fast processing!
We can use pipes to summarize data by year too:
# what was the annual air temperature average
year.sum <- harMet15.09.11 %>%
group_by(year) %>% # group by year
dplyr::summarize(mean(airt, na.rm=TRUE))
# what is the class of the output?
year.sum
## # A tibble: 3 x 2
## year `mean(airt, na.rm = TRUE)`
## <dbl> <dbl>
## 1 2009 7.63
## 2 2010 9.03
## 3 2011 8.75
# view structure of output
str(year.sum)
## tibble [3 × 2] (S3: tbl_df/tbl/data.frame)
## $ year : num [1:3] 2009 2010 2011
## $ mean(airt, na.rm = TRUE): num [1:3] 7.63 9.03 8.75
Other dplyr Functions
dplyr works based on a series of verb functions that allow us to manipulate
the data in different ways:
-
filter()&slice(): filter rows based on values in specified columns -
group-by(): group all data by a column -
arrange(): sort data by values in specified columns -
select()&rename(): view and work with data from only specified columns -
distinct(): view and work with only unique values from specified columns -
mutate()&transmute(): add new data to a data frame -
summarise(): calculate the specified summary statistics -
sample_n()&sample_frac(): return a random sample of rows
(List modified from the CRAN dplyr
package description. )
The syntax for all dplyr functions is similar:
- the first argument is the target
data_frame, - the subsequent arguments dictate what to do with that
data_frameand - the output is a new data frame.
Group by a Variable (or Two)
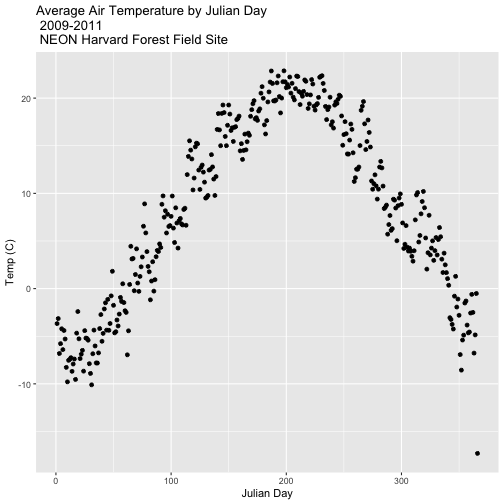
Our goal for this tutorial is to view drivers of annual phenology patterns.
Specifically, we want to explore daily average temperature throughout the year.
This means we need to calculate average temperature, for each day, across three
years. To do this we can use the group_by() function as we did earlier.
However this time, we can group by two variables: year and Julian Day (jd) as follows:
group_by(year, jd)
Let's begin by counting or tallying the total measurements per Julian day (or
year day) using the group_by() function and pipes.
harMet15.09.11 %>% # use the harMet15.09.11 data_frame
group_by(year, jd) %>% # group data by Year & Julian day
tally() # tally (count) observations per jd / year
## # A tibble: 1,096 x 3
## # Groups: year [3]
## year jd n
## <dbl> <int> <int>
## 1 2009 1 96
## 2 2009 2 96
## 3 2009 3 96
## 4 2009 4 96
## 5 2009 5 96
## 6 2009 6 96
## 7 2009 7 96
## 8 2009 8 96
## 9 2009 9 96
## 10 2009 10 96
## # … with 1,086 more rows
The output shows we have 96 values for each day. Is that what we expect?
24*4 # 24 hours/day * 4 15-min data points/hour
## [1] 96
Summarize by Group
We can use summarize() function to calculate a summary output value for each
"group" - in this case Julian day per year. Let's calculate the mean air
temperature for each Julian day per year. Note that we are still using
na.rm=TRUE to tell R to skip NA values.
harMet15.09.11 %>% # use the harMet15.09.11 data_frame
group_by(year, jd) %>% # group data by Year & Julian day
dplyr::summarize(mean_airt = mean(airt, na.rm = TRUE)) # mean airtemp per jd / year
## `summarise()` has grouped output by 'year'. You can override using the `.groups` argument.
## # A tibble: 1,096 x 3
## # Groups: year [3]
## year jd mean_airt
## <dbl> <int> <dbl>
## 1 2009 1 -15.1
## 2 2009 2 -9.14
## 3 2009 3 -5.54
## 4 2009 4 -6.35
## 5 2009 5 -2.41
## 6 2009 6 -4.92
## 7 2009 7 -2.59
## 8 2009 8 -3.23
## 9 2009 9 -9.92
## 10 2009 10 -11.1
## # … with 1,086 more rows
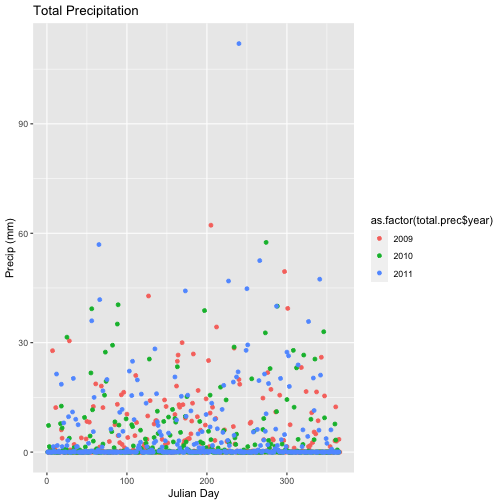
- Calculate total
precfor each Julian Day over the 3 years - name your data_frametotal.prec. - Create a plot of Daily Total Precipitation for 2009-2011.
- Add a title and x and y axis labels.
- If you use
qplotto create your plot, usecolour=as.factor(total.prec$year)to color the data points by year.
## `summarise()` has grouped output by 'year'. You can override using the `.groups` argument.
Mutate - Add data_frame Columns to dplyr Output
We can use the mutate() function of dplyr to add additional columns of
information to a data_frame. For instance, we added the year column
independently at the very beginning of the tutorial. However, we can add the
year using a dplyr pipe that also summarizes our data. To do this, we would
use the syntax:
mutate(year2 = year(datetime))
year2 is the name of the column that will be added to the output dplyr
data_frame.
harMet15.09.11 %>%
mutate(year2 = year(datetime)) %>%
group_by(year2, jd) %>%
dplyr::summarize(mean_airt = mean(airt, na.rm = TRUE))
## `summarise()` has grouped output by 'year2'. You can override using the `.groups` argument.
## # A tibble: 1,096 x 3
## # Groups: year2 [3]
## year2 jd mean_airt
## <dbl> <int> <dbl>
## 1 2009 1 -15.1
## 2 2009 2 -9.14
## 3 2009 3 -5.54
## 4 2009 4 -6.35
## 5 2009 5 -2.41
## 6 2009 6 -4.92
## 7 2009 7 -2.59
## 8 2009 8 -3.23
## 9 2009 9 -9.92
## 10 2009 10 -11.1
## # … with 1,086 more rows
Save dplyr Output as data_frame
We can save output from a dplyr pipe as a new R object to use for quick
plotting.
harTemp.daily.09.11<-harMet15.09.11 %>%
mutate(year2 = year(datetime)) %>%
group_by(year2, jd) %>%
dplyr::summarize(mean_airt = mean(airt, na.rm = TRUE))
## `summarise()` has grouped output by 'year2'. You can override using the `.groups` argument.
head(harTemp.daily.09.11)
## # A tibble: 6 x 3
## # Groups: year2 [1]
## year2 jd mean_airt
## <dbl> <int> <dbl>
## 1 2009 1 -15.1
## 2 2009 2 -9.14
## 3 2009 3 -5.54
## 4 2009 4 -6.35
## 5 2009 5 -2.41
## 6 2009 6 -4.92
Add Date-Time To dplyr Output
In the challenge above, we created a plot of daily precipitation data using
qplot. However, the x-axis ranged from 0-366 (Julian Days for the year). It
would have been easier to create a meaningful plot across all three years if we
had a continuous date variable in our data_frame representing the year and
date for each summary value.
We can add the the datetime column value to our data_frame by adding an
additional argument to the summarize() function. In this case, we will add the
first datetime value that R encounters when summarizing data by group as
follows:
datetime = first(datetime)
Our new summarize statement in our pipe will look like this:
summarize(mean_airt = mean(airt, na.rm = TRUE), datetime = first(datetime))
Let's try it!
# add in a datatime column
harTemp.daily.09.11 <- harMet15.09.11 %>%
mutate(year3 = year(datetime)) %>%
group_by(year3, jd) %>%
dplyr::summarize(mean_airt = mean(airt, na.rm = TRUE), datetime = first(datetime))
## `summarise()` has grouped output by 'year3'. You can override using the `.groups` argument.
# view str and head of data
str(harTemp.daily.09.11)
## tibble [1,096 × 4] (S3: grouped_df/tbl_df/tbl/data.frame)
## $ year3 : num [1:1096] 2009 2009 2009 2009 2009 ...
## $ jd : int [1:1096] 1 2 3 4 5 6 7 8 9 10 ...
## $ mean_airt: num [1:1096] -15.13 -9.14 -5.54 -6.35 -2.41 ...
## $ datetime : POSIXct[1:1096], format: "2009-01-01 00:15:00" ...
## - attr(*, "groups")= tibble [3 × 2] (S3: tbl_df/tbl/data.frame)
## ..$ year3: num [1:3] 2009 2010 2011
## ..$ .rows: list<int> [1:3]
## .. ..$ : int [1:366] 1 2 3 4 5 6 7 8 9 10 ...
## .. ..$ : int [1:365] 367 368 369 370 371 372 373 374 375 376 ...
## .. ..$ : int [1:365] 732 733 734 735 736 737 738 739 740 741 ...
## .. ..@ ptype: int(0)
## ..- attr(*, ".drop")= logi TRUE
head(harTemp.daily.09.11)
## # A tibble: 6 x 4
## # Groups: year3 [1]
## year3 jd mean_airt datetime
## <dbl> <int> <dbl> <dttm>
## 1 2009 1 -15.1 2009-01-01 00:15:00
## 2 2009 2 -9.14 2009-01-02 00:15:00
## 3 2009 3 -5.54 2009-01-03 00:15:00
## 4 2009 4 -6.35 2009-01-04 00:15:00
## 5 2009 5 -2.41 2009-01-05 00:15:00
## 6 2009 6 -4.92 2009-01-06 00:15:00
- Plot daily total precipitation from 2009-2011 as we did in the previous challenge. However this time, use the new syntax that you learned (mutate and summarize to add a datetime column to your output data_frame).
- Create a data_frame of the average monthly air temperature for 2009-2011.
Name the new data frame
harTemp.monthly.09.11. Plot your output.
## `summarise()` has grouped output by 'year'. You can override using the `.groups` argument.
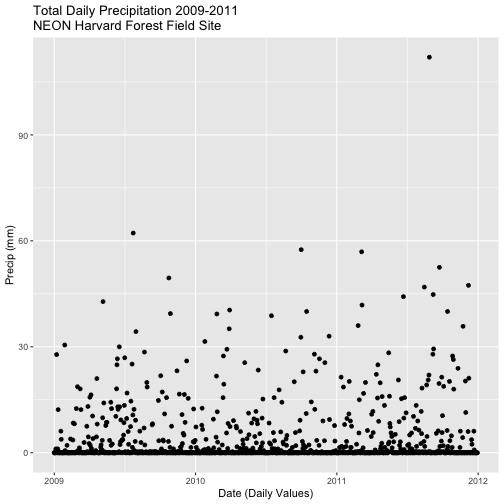
## `summarise()` has grouped output by 'month'. You can override using the `.groups` argument.
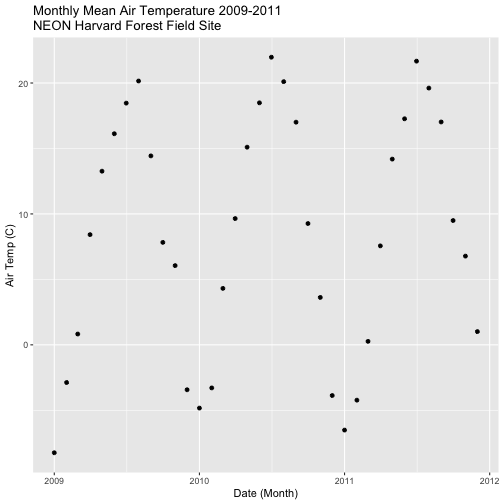
Get Lesson Code
Time Series 05: Plot Time Series with ggplot2 in R
Authors: Megan A. Jones, Marisa Guarinello, Courtney Soderberg, Leah A. Wasser
Last Updated: May 13, 2021
This tutorial uses ggplot2 to create customized plots of time
series data. We will learn how to adjust x- and y-axis ticks using the scales
package, how to add trend lines to a scatter plot and how to customize plot
labels, colors and overall plot appearance using ggthemes.
Learning Objectives
After completing this tutorial, you will be able to:
- Create basic time series plots using
ggplot()in R. - Explain the syntax of
ggplot()and know how to find out more about the package. - Plot data using scatter and bar plots.
Things You’ll Need To Complete This Tutorial
You will need the most current version of R and, preferably, RStudio loaded on your computer to complete this tutorial.
Install R Packages
-
lubridate:
install.packages("lubridate") -
ggplot2:
install.packages("ggplot2") -
scales:
install.packages("scales") -
gridExtra:
install.packages("gridExtra") -
ggthemes:
install.packages("ggthemes", dependencies = TRUE)
More on Packages in R – Adapted from Software Carpentry.
Download Data
NEON Teaching Data Subset: Meteorological Data for Harvard Forest
The data used in this lesson were collected at the National Ecological Observatory Network's Harvard Forest field site. These data are proxy data for what will be available for 30 years on the NEON data portal for the Harvard Forest and other field sites located across the United States.
Set Working Directory: This lesson assumes that you have set your working directory to the location of the downloaded and unzipped data subsets.
An overview of setting the working directory in R can be found here.
R Script & Challenge Code: NEON data lessons often contain challenges that reinforce learned skills. If available, the code for challenge solutions is found in the downloadable R script of the entire lesson, available in the footer of each lesson page.
Additional Resources
- Winston Chang's
Plotting Time Series Data
Plotting our data allows us to quickly see general patterns including
outlier points and trends. Plots are also a useful way to communicate the
results of our research. ggplot2 is a powerful R package that we use to
create customized, professional plots.
Load the Data
We will use the lubridate, ggplot2, scales and gridExtra packages in
this tutorial.
Our data subset will be the daily meteorology data for 2009-2011 for the NEON
Harvard Forest field site
(NEON-DS-Met-Time-Series/HARV/FisherTower-Met/Met_HARV_Daily_2009_2011.csv).
If this subset is not already loaded, please load it now.
# Remember it is good coding technique to add additional packages to the top of
# your script
library(lubridate) # for working with dates
library(ggplot2) # for creating graphs
library(scales) # to access breaks/formatting functions
library(gridExtra) # for arranging plots
# set working directory to ensure R can find the file we wish to import
wd <- "~/Git/data/"
# daily HARV met data, 2009-2011
harMetDaily.09.11 <- read.csv(
file=paste0(wd,"NEON-DS-Met-Time-Series/HARV/FisherTower-Met/Met_HARV_Daily_2009_2011.csv"),
stringsAsFactors = FALSE)
# covert date to Date class
harMetDaily.09.11$date <- as.Date(harMetDaily.09.11$date)
# monthly HARV temperature data, 2009-2011
harTemp.monthly.09.11<-read.csv(
file=paste0(wd,"NEON-DS-Met-Time-Series/HARV/FisherTower-Met/Temp_HARV_Monthly_09_11.csv"),
stringsAsFactors=FALSE
)
# datetime field is actually just a date
#str(harTemp.monthly.09.11)
# convert datetime from chr to date class & rename date for clarification
harTemp.monthly.09.11$date <- as.Date(harTemp.monthly.09.11$datetime)
Plot with qplot
We can use the qplot() function in the ggplot2 package to quickly plot a
variable such as air temperature (airt) across all three years of our daily
average time series data.
# plot air temp
qplot(x=date, y=airt,
data=harMetDaily.09.11, na.rm=TRUE,
main="Air temperature Harvard Forest\n 2009-2011",
xlab="Date", ylab="Temperature (°C)")
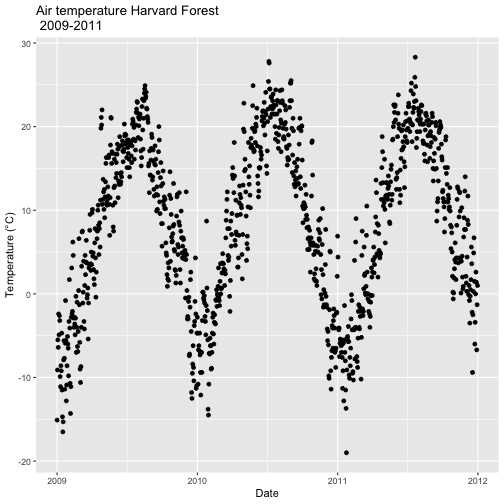
The resulting plot displays the pattern of air temperature increasing and
decreasing over three years. While qplot() is a quick way to plot data, our
ability to customize the output is limited.
Plot with ggplot
The ggplot() function within the ggplot2 package gives us more control
over plot appearance. However, to use ggplot we need to learn a slightly
different syntax. Three basic elements are needed for ggplot() to work:
- The data_frame: containing the variables that we wish to plot,
-
aes(aesthetics): which denotes which variables will map to the x-, y- (and other) axes, -
geom_XXXX(geometry): which defines the data's graphical representation (e.g. points (geom_point), bars (geom_bar), lines (geom_line), etc).
The syntax begins with the base statement that includes the data_frame
(harMetDaily.09.11) and associated x (date) and y (airt) variables to be
plotted:
ggplot(harMetDaily.09.11, aes(date, airt))
To successfully plot, the last piece that is needed is the geometry type. In
this case, we want to create a scatterplot so we can add + geom_point().
Let's create an air temperature scatterplot.
# plot Air Temperature Data across 2009-2011 using daily data
ggplot(harMetDaily.09.11, aes(date, airt)) +
geom_point(na.rm=TRUE)
Customize A Scatterplot
We can customize our plot in many ways. For instance, we can change the size and
color of the points using size=, shape pch=, and color= in the geom_point
element.
geom_point(na.rm=TRUE, color="blue", size=1)
# plot Air Temperature Data across 2009-2011 using daily data
ggplot(harMetDaily.09.11, aes(date, airt)) +
geom_point(na.rm=TRUE, color="blue", size=3, pch=18)
Modify Title & Axis Labels
We can modify plot attributes by adding elements using the + symbol.
For example, we can add a title by using + ggtitle="TEXT", and axis
labels using + xlab("TEXT") + ylab("TEXT").
# plot Air Temperature Data across 2009-2011 using daily data
ggplot(harMetDaily.09.11, aes(date, airt)) +
geom_point(na.rm=TRUE, color="blue", size=1) +
ggtitle("Air Temperature 2009-2011\n NEON Harvard Forest Field Site") +
xlab("Date") + ylab("Air Temperature (C)")
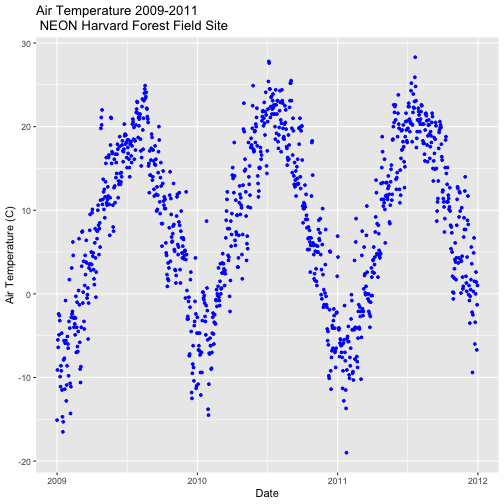
Name Plot Objects
We can create a ggplot object by assigning our plot to an object name.
When we do this, the plot will not render automatically. To render the plot, we
need to call it in the code.
Assigning plots to an R object allows us to effectively add on to,
and modify the plot later. Let's create a new plot and call it AirTempDaily.
# plot Air Temperature Data across 2009-2011 using daily data
AirTempDaily <- ggplot(harMetDaily.09.11, aes(date, airt)) +
geom_point(na.rm=TRUE, color="purple", size=1) +
ggtitle("Air Temperature\n 2009-2011\n NEON Harvard Forest") +
xlab("Date") + ylab("Air Temperature (C)")
# render the plot
AirTempDaily
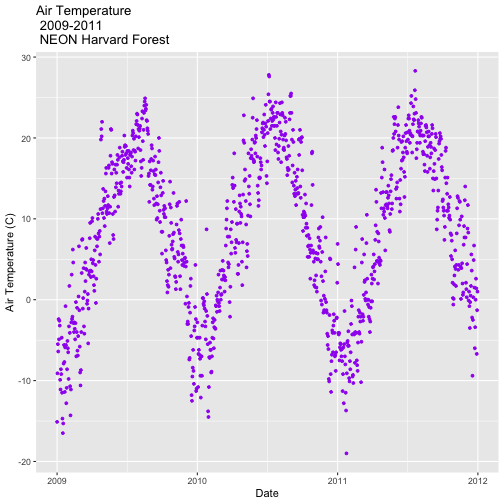
Format Dates in Axis Labels
We can adjust the date display format (e.g. 2009-07 vs. Jul 09) and the number
of major and minor ticks for axis date values using scale_x_date. Let's
format the axis ticks so they read "month year" (%b %y). To do this, we will
use the syntax:
scale_x_date(labels=date_format("%b %y")
Rather than re-coding the entire plot, we can add the scale_x_date element
to the plot object AirTempDaily that we just created.
# format x-axis: dates
AirTempDailyb <- AirTempDaily +
(scale_x_date(labels=date_format("%b %y")))
AirTempDailyb
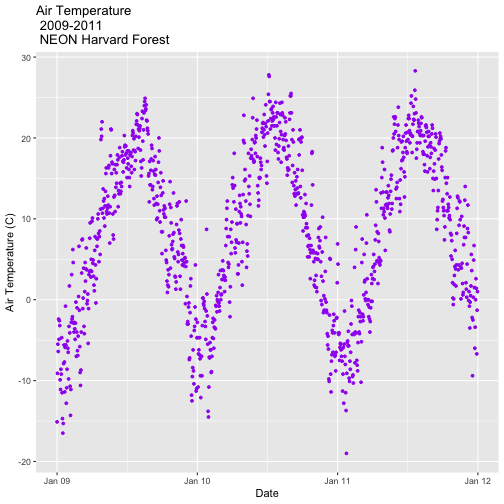
Adjust Date Ticks
We can adjust the date ticks too. In this instance, having 1 tick per year may
be enough. If we have the scales package loaded, we can use
breaks=date_breaks("1 year") within the scale_x_date element to create
a tick for every year. We can adjust this as needed (e.g. 10 days, 30 days, 1
month).
From R HELP (
?date_breaks):widthan interval specification, one of "sec", "min", "hour", "day", "week", "month", "year". Can be by an integer and a space, or followed by "s".
# format x-axis: dates
AirTempDaily_6mo <- AirTempDaily +
(scale_x_date(breaks=date_breaks("6 months"),
labels=date_format("%b %y")))
AirTempDaily_6mo
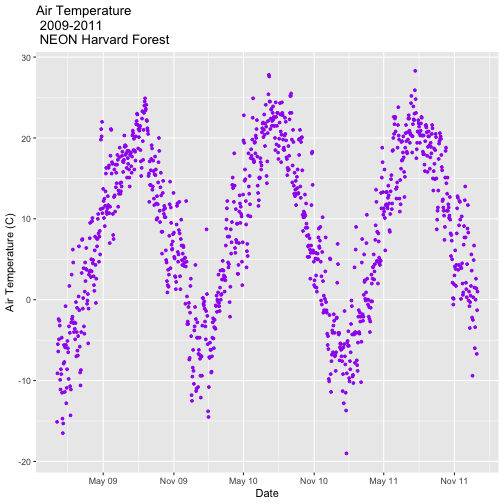
# format x-axis: dates
AirTempDaily_1y <- AirTempDaily +
(scale_x_date(breaks=date_breaks("1 year"),
labels=date_format("%b %y")))
AirTempDaily_1y
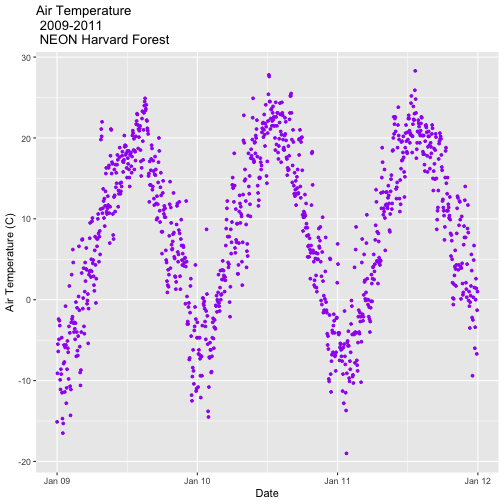
ggplot - Subset by Time
Sometimes we want to scale the x- or y-axis to a particular time subset without
subsetting the entire data_frame. To do this, we can define start and end
times. We can then define the limits in the scale_x_date object as
follows:
scale_x_date(limits=start.end) +
# Define Start and end times for the subset as R objects that are the time class
startTime <- as.Date("2011-01-01")
endTime <- as.Date("2012-01-01")
# create a start and end time R object
start.end <- c(startTime,endTime)
start.end
## [1] "2011-01-01" "2012-01-01"
# View data for 2011 only
# We will replot the entire plot as the title has now changed.
AirTempDaily_2011 <- ggplot(harMetDaily.09.11, aes(date, airt)) +
geom_point(na.rm=TRUE, color="purple", size=1) +
ggtitle("Air Temperature\n 2011\n NEON Harvard Forest") +
xlab("Date") + ylab("Air Temperature (C)")+
(scale_x_date(limits=start.end,
breaks=date_breaks("1 year"),
labels=date_format("%b %y")))
AirTempDaily_2011
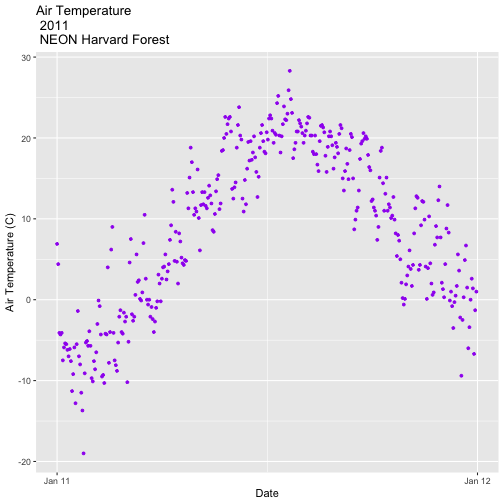
ggplot() Themes
We can use the theme() element to adjust figure elements.
There are some nice pre-defined themes that we can use as a starting place.
# Apply a black and white stock ggplot theme
AirTempDaily_bw<-AirTempDaily_1y +
theme_bw()
AirTempDaily_bw
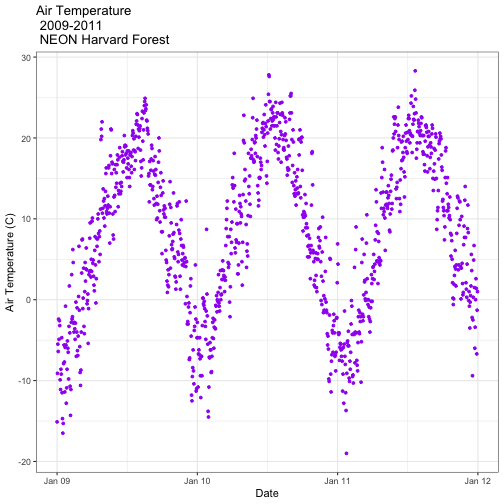
Using the theme_bw() we now have a white background rather than grey.
Import New Themes Bonus Topic
There are externally developed themes built by the R community that are worth
mentioning. Feel free to experiment with the code below to install ggthemes.
# install additional themes
# install.packages('ggthemes', dependencies = TRUE)
library(ggthemes)
AirTempDaily_economist<-AirTempDaily_1y +
theme_economist()
AirTempDaily_economist
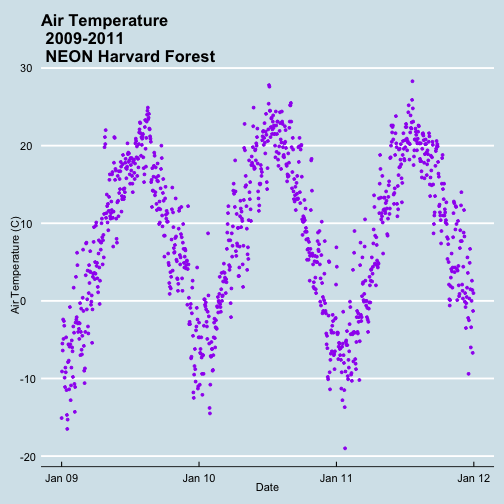
AirTempDaily_strata<-AirTempDaily_1y +
theme_stata()
AirTempDaily_strata
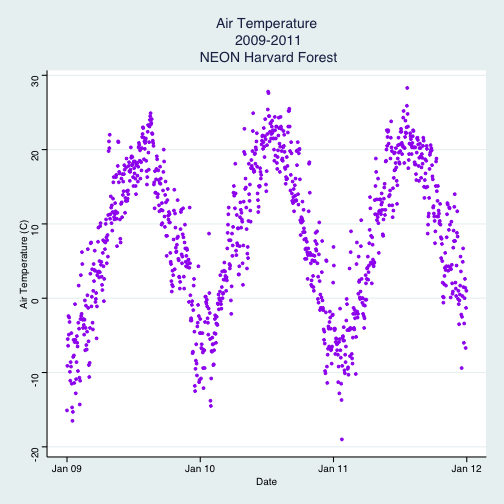
More on Themes
Hadley Wickham's documentation.
A list of themes loaded in the ggthemes library is found here.
Customize ggplot Themes
We can customize theme elements manually too. Let's customize the font size and style.
# format x axis with dates
AirTempDaily_custom<-AirTempDaily_1y +
# theme(plot.title) allows to format the Title separately from other text
theme(plot.title = element_text(lineheight=.8, face="bold",size = 20)) +
# theme(text) will format all text that isn't specifically formatted elsewhere
theme(text = element_text(size=18))
AirTempDaily_custom
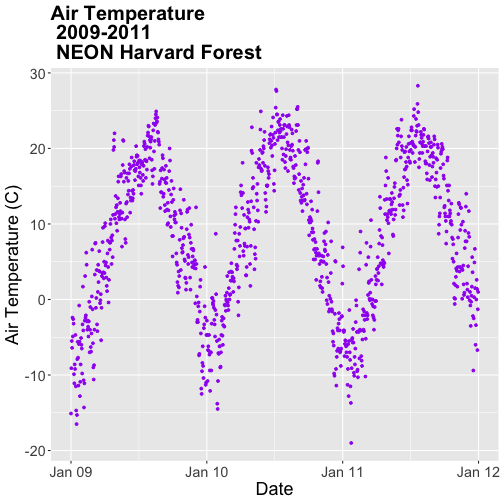
- Format the dates on the x-axis:
Month-Year. - Create a plot object called
PrecipDaily. - Be sure to add an appropriate title in addition to x and y axis labels.
- Increase the font size of the plot text and adjust the number of ticks on the x-axis.
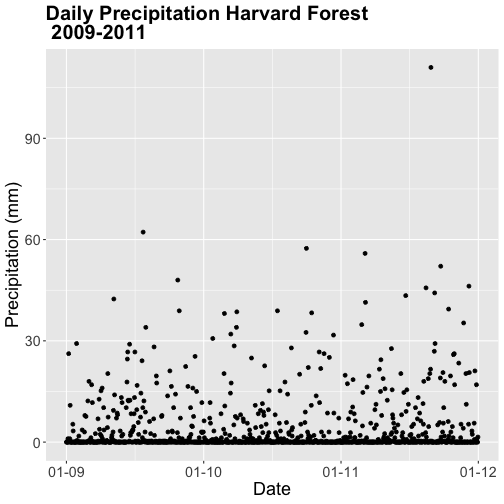
Bar Plots with ggplot
We can use ggplot to create bar plots too. Let's create a bar plot of total
daily precipitation next. A bar plot might be a better way to represent a total
daily value. To create a bar plot, we change the geom element from
geom_point() to geom_bar().
The default setting for a ggplot bar plot - geom_bar() - is a histogram
designated by stat="bin". However, in this case, we want to plot actual
precipitation values. We can use geom_bar(stat="identity") to force ggplot to
plot actual values.
# plot precip
PrecipDailyBarA <- ggplot(harMetDaily.09.11, aes(date, prec)) +
geom_bar(stat="identity", na.rm = TRUE) +
ggtitle("Daily Precipitation\n Harvard Forest") +
xlab("Date") + ylab("Precipitation (mm)") +
scale_x_date(labels=date_format ("%b %y"), breaks=date_breaks("1 year")) +
theme(plot.title = element_text(lineheight=.8, face="bold", size = 20)) +
theme(text = element_text(size=18))
PrecipDailyBarA
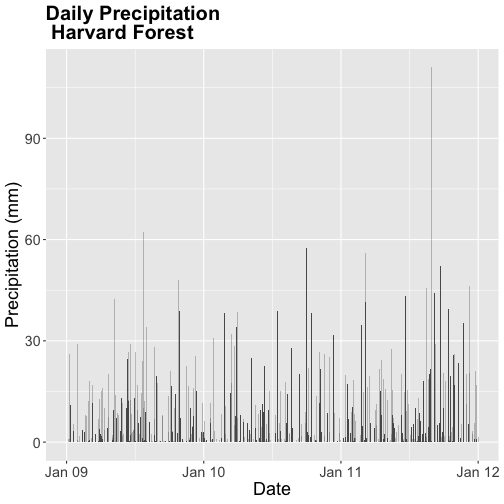
Note that some of the bars in the resulting plot appear grey rather than black. This is because R will do it's best to adjust colors of bars that are closely spaced to improve readability. If we zoom into the plot, all of the bars are black.
- a descriptive title and axis labels,
- breaks every 4 months, and
- x-axis labels as only the full month (spelled out).
HINT: you will need to rebuild the precipitation plot as you will have to
specify a new scale_x_data() element.
Bonus: Style your plot with a ggtheme of choice.
## Warning: Removed 729 rows containing missing values (position_stack).
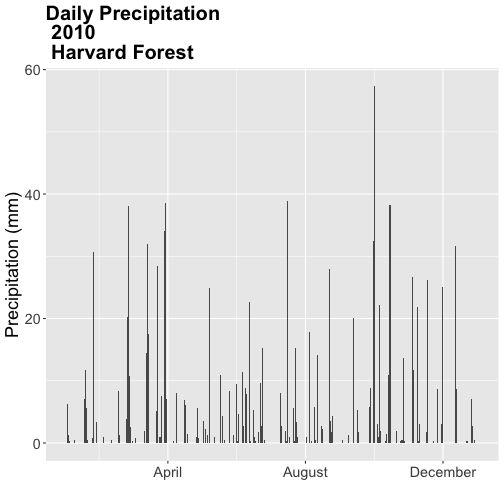
Color
We can change the bar fill color by within the
geom_bar(colour="blue") element. We can also specify a separate fill and line
color using fill= and line=. Colors can be specified by name (e.g.,
"blue") or hexadecimal color codes (e.g, #FF9999).
There are many color cheatsheets out there to help with color selection!
# specifying color by name
PrecipDailyBarB <- PrecipDailyBarA+
geom_bar(stat="identity", colour="darkblue")
PrecipDailyBarB
Figures with Lines
We can create line plots too using ggplot. To do this, we use geom_line()
instead of bar or point.
AirTempDaily_line <- ggplot(harMetDaily.09.11, aes(date, airt)) +
geom_line(na.rm=TRUE) +
ggtitle("Air Temperature Harvard Forest\n 2009-2011") +
xlab("Date") + ylab("Air Temperature (C)") +
scale_x_date(labels=date_format ("%b %y")) +
theme(plot.title = element_text(lineheight=.8, face="bold",
size = 20)) +
theme(text = element_text(size=18))
AirTempDaily_line
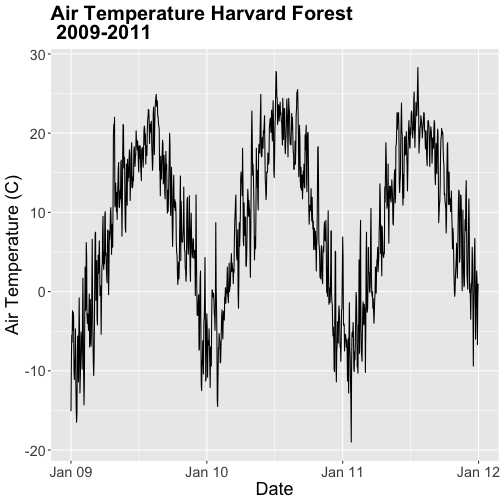
Note that lines may not be the best way to represent air temperature data given lines suggest that the connecting points are directly related. It is important to consider what type of plot best represents the type of data that you are presenting.
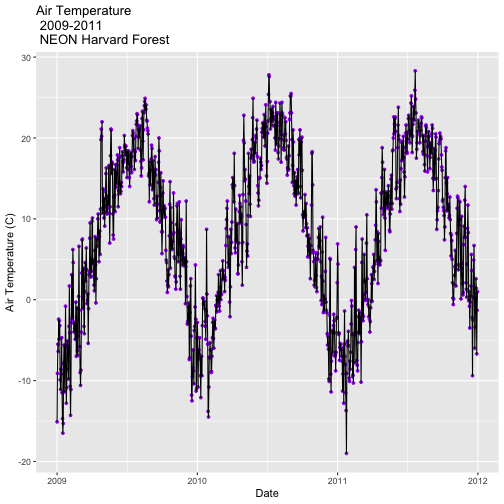
Trend Lines
We can add a trend line, which is a statistical transformation of our data to
represent general patterns, using stat_smooth(). stat_smooth() requires a
statistical method as follows:
- For data with < 1000 observations: the default model is a loess model (a non-parametric regression model)
- For data with > 1,000 observations: the default model is a GAM (a general additive model)
- A specific model/method can also be specified: for example, a linear regression (
method="lm").
For this tutorial, we will use the default trend line model. The gam method will
be used with given we have 1,095 measurements.
# adding on a trend lin using loess
AirTempDaily_trend <- AirTempDaily + stat_smooth(colour="green")
AirTempDaily_trend
## `geom_smooth()` using method = 'gam' and formula 'y ~ s(x, bs = "cs")'
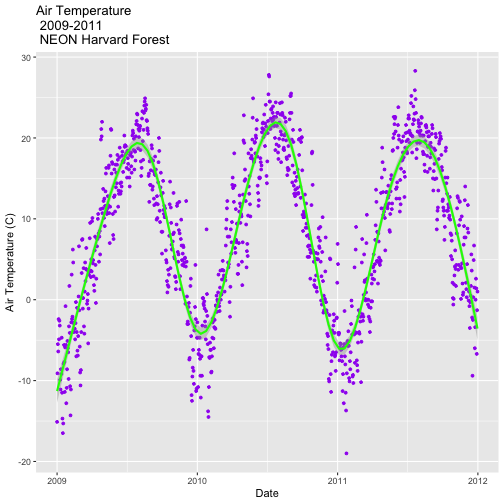
Create a bar plot of total daily precipitation. Add a:
- Trend line for total daily precipitation.
- Make the bars purple (or your favorite color!).
- Make the trend line grey (or your other favorite color).
- Adjust the tick spacing and format the dates to appear as "Jan 2009".
- Render the title in italics.
## `geom_smooth()` using formula 'y ~ x'
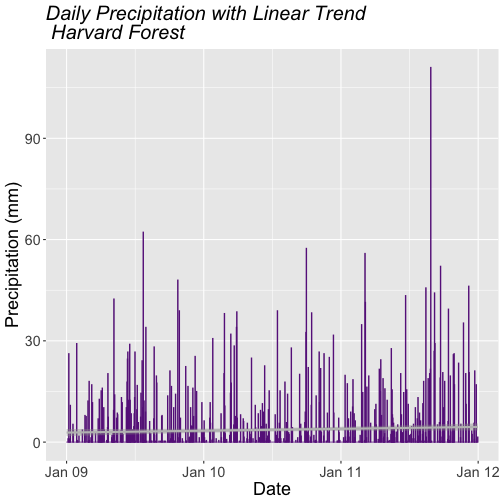
Plot the monthly air temperature across 2009-2011 using the
harTemp.monthly.09.11 data_frame. Name your plot "AirTempMonthly". Be sure to
label axes and adjust the plot ticks as you see fit.
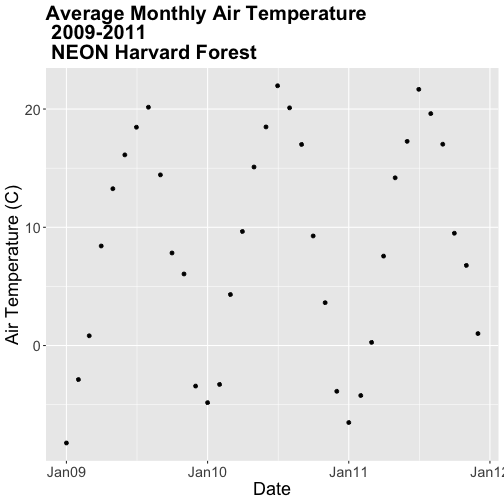
Display Multiple Figures in Same Panel
It is often useful to arrange plots in a panel rather than displaying them
individually. In base R, we use par(mfrow=()) to accomplish this. However
the grid.arrange() function from the gridExtra package provides a more
efficient approach!
grid.arrange requires 2 things:
- the names of the plots that you wish to render in the panel.
- the number of columns (
ncol).
grid.arrange(plotOne, plotTwo, ncol=1)
Let's plot AirTempMonthly and AirTempDaily on top of each other. To do this,
we'll specify one column.
# note - be sure library(gridExtra) is loaded!
# stack plots in one column
grid.arrange(AirTempDaily, AirTempMonthly, ncol=1)

Plot AirTempMonthly and AirTempDaily next to each other rather than stacked
on top of each other.
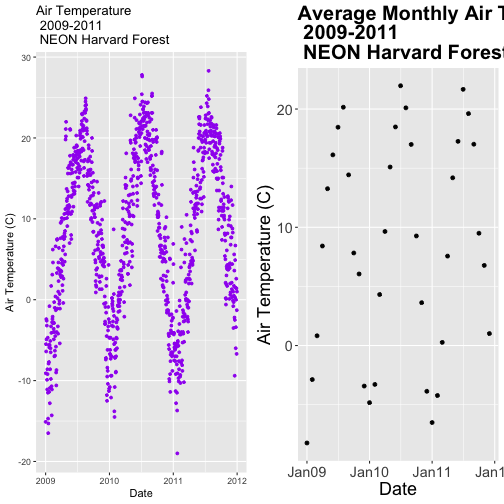
Additional ggplot2 Resources
In this tutorial, we've covered the basics of ggplot. There are many great
resources the cover refining ggplot figures. A few are below:
- ggplot2 Cheatsheet from Zev Ross: ggplot2 Cheatsheet
- ggplot2 documentation index: ggplot2 Documentation
Get Lesson Code
Time Series 06: Create Plots with Multiple Panels, Grouped by Time Using ggplot Facets
Authors: Megan A. Jones, Marisa Guarinello, Courtney Soderberg, Leah Wasser
Last Updated: Apr 10, 2025
This tutorial covers how to plot subsetted time series data using the facets()
function (e.g., plot by season) and to plot time series data with NDVI.
Learning Objectives
After completing this tutorial, you will be able to:
- Use
facets()in theggplot2package. - Combine different types of data into one plot layout.
Things You’ll Need To Complete This Tutorial
You will need the most current version of R and, preferably, RStudio loaded on your computer to complete this tutorial.
Install R Packages
-
ggplot2:
install.packages("ggplot2") -
scales:
install.packages("scales") -
gridExtra:
install.packages("gridExtra") -
grid:
install.packages("grid") -
dplyr:
install.packages("dplyr") -
reshape2:
install.packages("reshape2") -
zoo:
install.packages("zoo")
More on Packages in R – Adapted from Software Carpentry.
Download Data
NEON Teaching Data Subset: Meteorological Data for Harvard Forest
The data used in this lesson were collected at the National Ecological Observatory Network's Harvard Forest field site. These data are proxy data for what will be available for 30 years on the NEON data portal for the Harvard Forest and other field sites located across the United States.
Set Working Directory: This lesson assumes that you have set your working directory to the location of the downloaded and unzipped data subsets.
An overview of setting the working directory in R can be found here.
R Script & Challenge Code: NEON data lessons often contain challenges that reinforce learned skills. If available, the code for challenge solutions is found in the downloadable R script of the entire lesson, available in the footer of each lesson page.
Recommended Tutorials
This tutorial uses both dplyr and ggplot2. If you are new to either of these
R packages, we recommend the following NEON Data Skills tutorials before
working through this one.
Plot Data Subsets Using Facets
In this tutorial we will learn how to create a panel of individual plots - known
as facets in ggplot2. Each plot represents a particular data_frame
time-series subset, for example a year or a season.
Load the Data
We will use the daily micro-meteorology data for 2009-2011 from the Harvard
Forest. If you do not have this data loaded into an R data_frame, please
load them and convert date-time columns to a date-time class now.
# Remember it is good coding technique to add additional libraries to the top of
# your script
library(lubridate) # for working with dates
library(ggplot2) # for creating graphs
library(scales) # to access breaks/formatting functions
library(gridExtra) # for arranging plots
library(grid) # for arranging plots
library(dplyr) # for subsetting by season
# set working directory to ensure R can find the file we wish to import
wd <- "~/Git/data/"
# daily HARV met data, 2009-2011
harMetDaily.09.11 <- read.csv(
file=paste0(wd,"NEON-DS-Met-Time-Series/HARV/FisherTower-Met/Met_HARV_Daily_2009_2011.csv"),
stringsAsFactors = FALSE
)
# covert date to Date class
harMetDaily.09.11$date <- as.Date(harMetDaily.09.11$date)
ggplot2 Facets
Facets allow us to plot subsets of data in one cleanly organized panel. We use
facet_grid() to create a plot of a particular variable subsetted by a
particular group.
Let's plot air temperature as we did previously. We will name the ggplot
object AirTempDaily.
AirTempDaily <- ggplot(harMetDaily.09.11, aes(date, airt)) +
geom_point() +
ggtitle("Daily Air Temperature\n NEON Harvard Forest\n 2009-2011") +
xlab("Date") + ylab("Temperature (C)") +
scale_x_date(labels=date_format ("%m-%y"))+
theme(plot.title = element_text(lineheight=.8, face="bold",
size = 20)) +
theme(text = element_text(size=18))
AirTempDaily
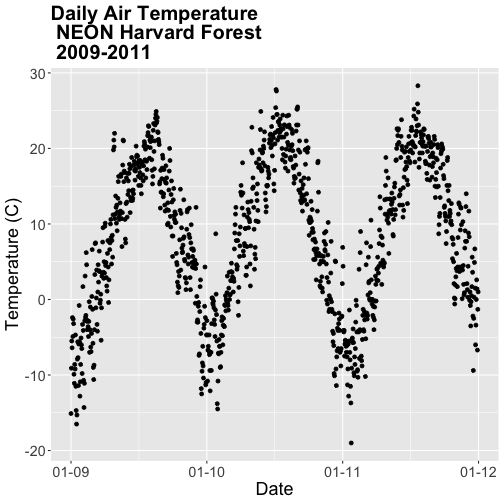
This plot tells us a lot about the annual increase and decrease of temperature at the NEON Harvard Forest field site. However, what if we want to plot each year's worth of data individually?
We can use the facet() element in ggplot to create facets or a panel of
plots that are grouped by a particular category or time period. To create a
plot for each year, we will first need a year column in our data to use as a
subset factor. We created a year column using the year function in the
lubridate package in the
Subset and Manipulate Time Series Data with dplyr tutorial.
# add year column to daily values
harMetDaily.09.11$year <- year(harMetDaily.09.11$date)
# view year column head and tail
head(harMetDaily.09.11$year)
## [1] 2009 2009 2009 2009 2009 2009
tail(harMetDaily.09.11$year)
## [1] 2011 2011 2011 2011 2011 2011
Facet by Year
Once we have a column that can be used to group or subset our data, we can create a faceted plot - plotting each year's worth of data in an individual, labelled panel.
# run this code to plot the same plot as before but with one plot per season
AirTempDaily + facet_grid(. ~ year)
## Error: At least one layer must contain all faceting variables: `year`.
## * Plot is missing `year`
## * Layer 1 is missing `year`

Oops - what happened? The plot did not render because we added the year column
after creating the ggplot object AirTempDaily. Let's rerun the plotting code
to ensure our newly added column is recognized.
AirTempDaily <- ggplot(harMetDaily.09.11, aes(date, airt)) +
geom_point() +
ggtitle("Daily Air Temperature\n NEON Harvard Forest") +
xlab("Date") + ylab("Temperature (C)") +
scale_x_date(labels=date_format ("%m-%y"))+
theme(plot.title = element_text(lineheight=.8, face="bold",
size = 20)) +
theme(text = element_text(size=18))
# facet plot by year
AirTempDaily + facet_grid(. ~ year)
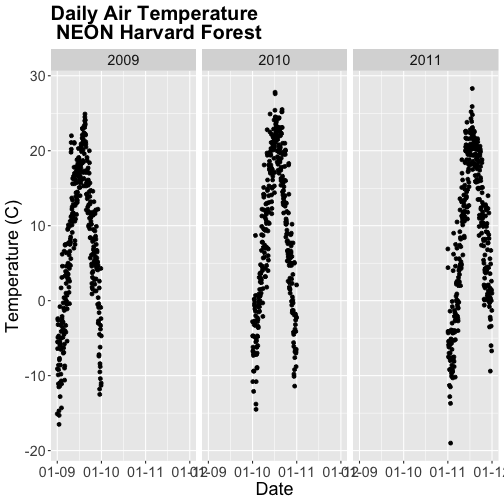
The faceted plot is interesting, however the x-axis on each plot is formatted as: month-day-year starting in 2009 and ending in 2011. This means that the data for 2009 is on the left end of the x-axis and the data for 2011 is on the right end of the x-axis of the 2011 plot.
Our plots would be easier to visually compare if the days were formatted in
Julian or year days rather than date. We have Julian days stored in our
data_frame (harMetDaily.09.11) as jd.
AirTempDaily_jd <- ggplot(harMetDaily.09.11, aes(jd, airt)) +
geom_point() +
ggtitle("Air Temperature\n NEON Harvard Forest Field Site") +
xlab("Julian Day") + ylab("Temperature (C)") +
theme(plot.title = element_text(lineheight=.8, face="bold",
size = 20)) +
theme(text = element_text(size=18))
# create faceted panel
AirTempDaily_jd + facet_grid(. ~ year)
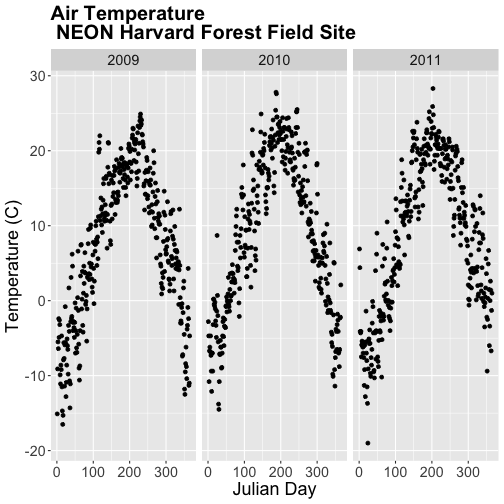
Using Julian day, our plots are easier to visually compare. Arranging our plots this way, side by side, allows us to quickly scan for differences along the y-axis. Notice any differences in min vs max air temperature across the three years?
Arrange Facets
We can rearrange the facets in different ways, too.
# move labels to the RIGHT and stack all plots
AirTempDaily_jd + facet_grid(year ~ .)
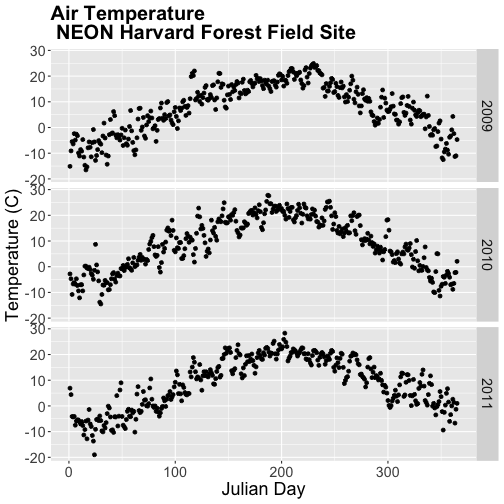
If we use facet_wrap we can specify the number of columns.
# display in two columns
AirTempDaily_jd + facet_wrap(~year, ncol = 2)
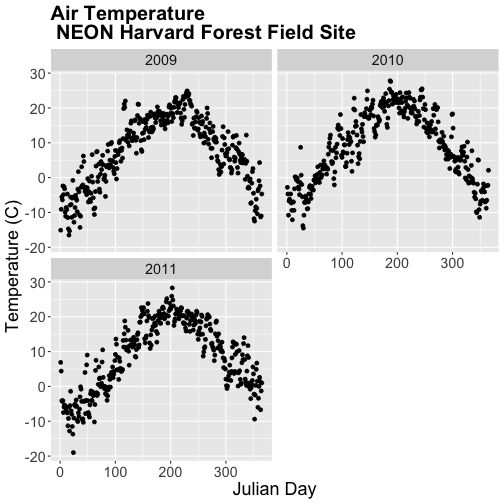
Graph Two Variables on One Plot
Next, let's explore the relationship between two variables - air temperature and soil temperature. We might expect soil temperature to fluctuate with changes in air temperature over time.
We will use ggplot() to plot airt and s10t (soil temperature 10 cm below
the ground).
airSoilTemp_Plot <- ggplot(harMetDaily.09.11, aes(airt, s10t)) +
geom_point() +
ggtitle("Air vs. Soil Temperature\n NEON Harvard Forest Field Site\n 2009-2011") +
xlab("Air Temperature (C)") + ylab("Soil Temperature (C)") +
theme(plot.title = element_text(lineheight=.8, face="bold",
size = 20)) +
theme(text = element_text(size=18))
airSoilTemp_Plot
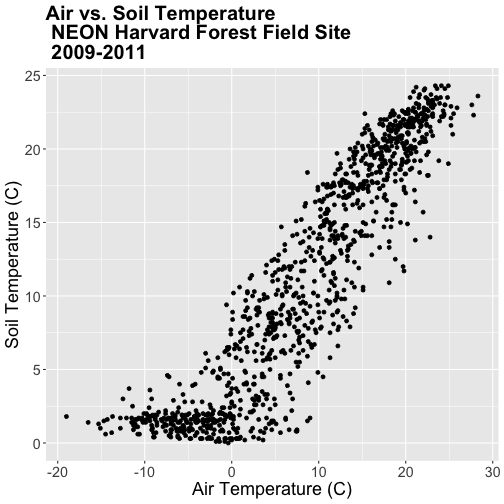
The plot above suggests a relationship between the air and soil temperature as we might expect. However, it clumps all three years worth of data into one plot.
Let's create a stacked faceted plot of air vs. soil temperature grouped by year.
Lucky for us, we can do this quickly with one line of code while reusing the plot we created above.
# create faceted panel
airSoilTemp_Plot + facet_grid(year ~ .)
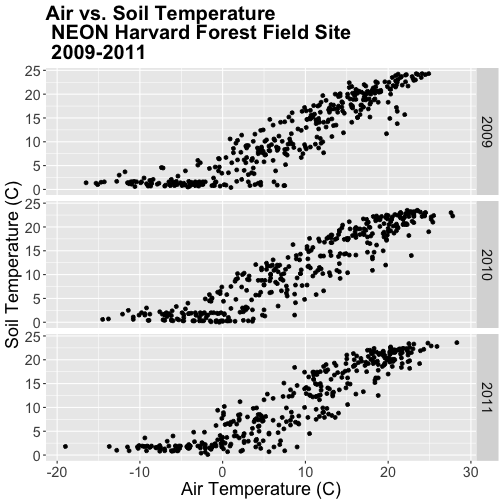
Have a close look at the data. Are there any noticeable min/max temperature differences between the three years?
Create a faceted plot of air temperature vs soil temperature by month rather than year.
HINT: To create this plot, you will want to add a month column to our
data_frame. We can use lubridate month in the same way we used year to add
a year column.
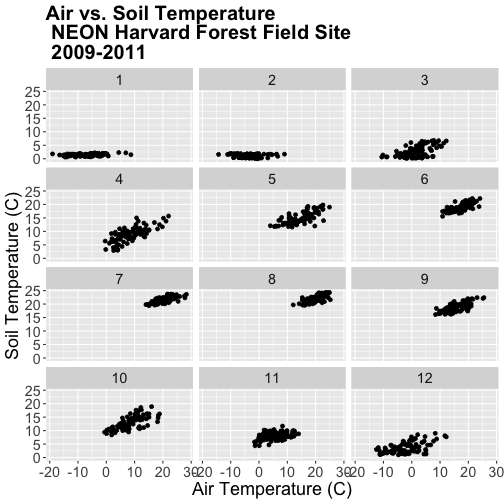
Faceted Plots & Categorical Groups
In the challenge above, we grouped our data by month - specified by a numeric value between 1 (January) and 12 (December). However, what if we wanted to organize our plots using a categorical (character) group such as month name? Let's do that next.
If we want to group our data by month name, we first need to create a month
name column in our data_frame. We can create this column using the following
syntax:
format(harMetDaily.09.11$date,"%B"),
which tells R to extract the month name (%B) from the date field.
# add text month name column
harMetDaily.09.11$month_name <- format(harMetDaily.09.11$date,"%B")
# view head and tail
head(harMetDaily.09.11$month_name)
## [1] "January" "January" "January" "January" "January" "January"
tail(harMetDaily.09.11$month_name)
## [1] "December" "December" "December" "December" "December" "December"
# recreate plot
airSoilTemp_Plot <- ggplot(harMetDaily.09.11, aes(airt, s10t)) +
geom_point() +
ggtitle("Air vs. Soil Temperature \n NEON Harvard Forest Field Site\n 2009-2011") +
xlab("Air Temperature (C)") + ylab("Soil Temperature (C)") +
theme(plot.title = element_text(lineheight=.8, face="bold",
size = 20)) +
theme(text = element_text(size=18))
# create faceted panel
airSoilTemp_Plot + facet_wrap(~month_name, nc=3)
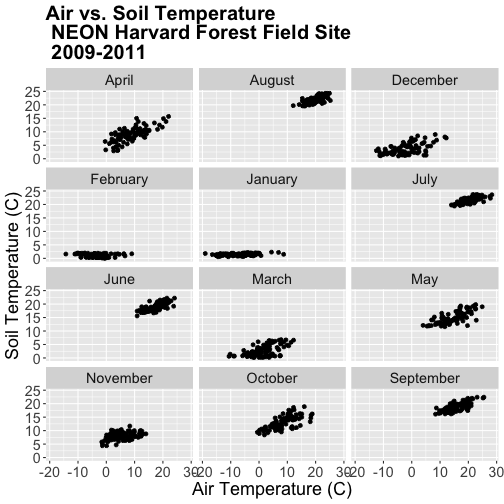
Great! We've created a nice set of plots by month. However, how are the plots
ordered? It looks like R is ordering things alphabetically, yet we know
that months are ordinal not character strings. To account for order, we can
reassign the month_name field to a factor. This will allow us to specify
an order to each factor "level" (each month is a level).
The syntax for this operation is
- Turn field into a factor:
factor(fieldName). - Designate the
levelsusing a listc(level1, level2, level3).
In our case, each level will be a month.
# order the factors
harMetDaily.09.11$month_name = factor(harMetDaily.09.11$month_name,
levels=c('January','February','March',
'April','May','June','July',
'August','September','October',
'November','December'))
Once we have specified the factor column and its associated levels, we can plot
again. Remember, that because we have modified a column in our data_frame, we
need to rerun our ggplot code.
# recreate plot
airSoilTemp_Plot <- ggplot(harMetDaily.09.11, aes(airt, s10t)) +
geom_point() +
ggtitle("Air vs. Soil Temperature \n NEON Harvard Forest Field Site\n 2009-2011") +
xlab("Air Temperature (C)") + ylab("Soil Temperature (C)") +
theme(plot.title = element_text(lineheight=.8, face="bold",
size = 20)) +
theme(text = element_text(size=18))
# create faceted panel
airSoilTemp_Plot + facet_wrap(~month_name, nc=3)
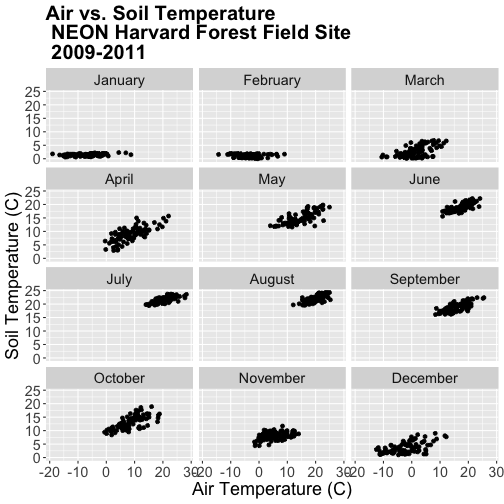
Subset by Season - Advanced Topic
Sometimes we want to group data by custom time periods. For example, we might want to group by season. However, the definition of various seasons may vary by region which means we need to manually define each time period.
In the next coding section, we will add a season column to our data using a manually defined query. Our field site is Harvard Forest (Massachusetts), located in the northeastern portion of the United States. Based on the weather of this region, we can divide the year into 4 seasons as follows:
- Winter: December - February
- Spring: March - May
- Summer: June - August
- Fall: September - November
In order to subset the data by season we will use the dplyr package. We
can use the numeric month column that we added to our data earlier in this
tutorial.
# add month to data_frame - note we already performed this step above.
harMetDaily.09.11$month <- month(harMetDaily.09.11$date)
# view head and tail of column
head(harMetDaily.09.11$month)
## [1] 1 1 1 1 1 1
tail(harMetDaily.09.11$month)
## [1] 12 12 12 12 12 12
We can use mutate() and a set of ifelse statements to create a new
categorical variable called season by grouping three months together.
Within dplyr %in% is short-hand for "contained within". So the syntax
ifelse(month %in% c(12, 1, 2), "Winter",
can be read as "if the month column value is 12 or 1 or 2, then assign the
value "Winter"".
Our ifelse statement ends with
ifelse(month %in% c(9, 10, 11), "Fall", "Error")
which we can translate this as "if the month column value is 9 or 10 or 11,
then assign the value "Winter"."
The last portion , "Error" tells R that if a month column value does not
fall within any of the criteria laid out in previous ifelse statements,
assign the column the value of "Error".
harMetDaily.09.11 <- harMetDaily.09.11 %>%
mutate(season =
ifelse(month %in% c(12, 1, 2), "Winter",
ifelse(month %in% c(3, 4, 5), "Spring",
ifelse(month %in% c(6, 7, 8), "Summer",
ifelse(month %in% c(9, 10, 11), "Fall", "Error")))))
# check to see if this worked
head(harMetDaily.09.11$month)
## [1] 1 1 1 1 1 1
head(harMetDaily.09.11$season)
## [1] "Winter" "Winter" "Winter" "Winter" "Winter" "Winter"
tail(harMetDaily.09.11$month)
## [1] 12 12 12 12 12 12
tail(harMetDaily.09.11$season)
## [1] "Winter" "Winter" "Winter" "Winter" "Winter" "Winter"
Now that we have a season column, we can plot our data by season!
# recreate plot
airSoilTemp_Plot <- ggplot(harMetDaily.09.11, aes(airt, s10t)) +
geom_point() +
ggtitle("Air vs. Soil Temperature\n 2009-2011\n NEON Harvard Forest Field Site") +
xlab("Air Temperature (C)") + ylab("Soil Temperature (C)") +
theme(plot.title = element_text(lineheight=.8, face="bold",
size = 20)) +
theme(text = element_text(size=18))
# run this code to plot the same plot as before but with one plot per season
airSoilTemp_Plot + facet_grid(. ~ season)
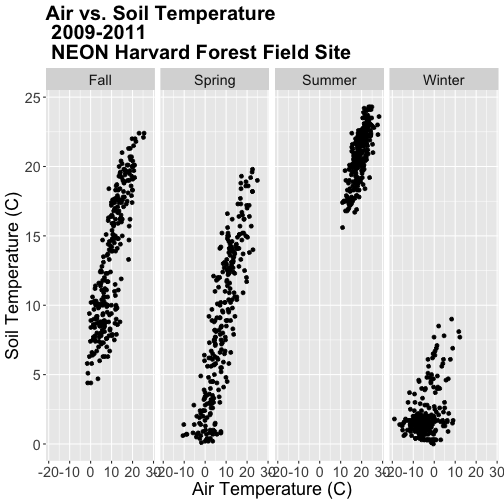
Note, that once again, we re-ran our ggplot code to make sure our new column
is recognized by R. We can experiment with various facet layouts next.
# for a landscape orientation of the plots we change the order of arguments in
# facet_grid():
airSoilTemp_Plot + facet_grid(season ~ .)
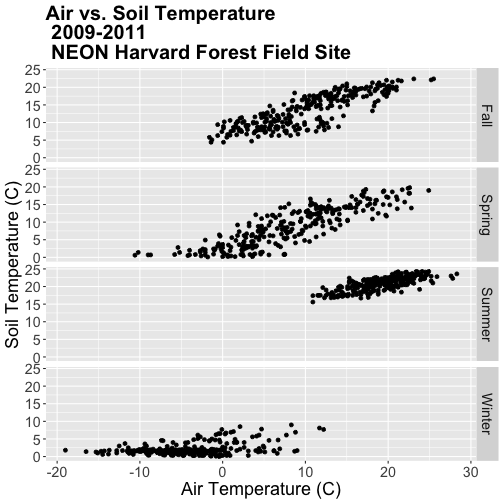
Once again, R is arranging the plots in an alphabetical order not an order relevant to the data.
-
Create a factor class season variable by converting the season column that we just created to a factor, then organize the seasons chronologically as follows: Winter, Spring, Summer, Fall.
-
Create a new faceted plot that is 2 x 2 (2 columns of plots). HINT: One can neatly plot multiple variables using facets as follows:
facet_grid(variable1 ~ variable2). -
Create a plot of air vs soil temperature grouped by year and season.
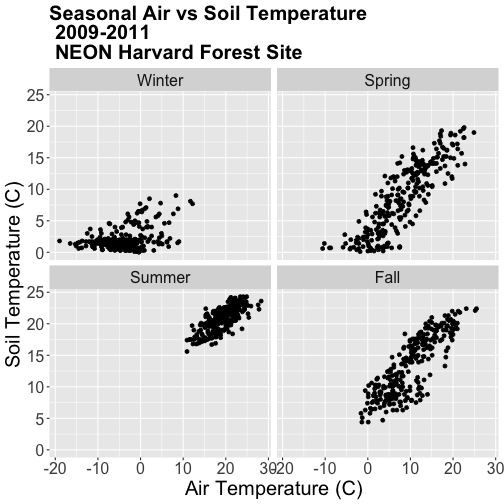
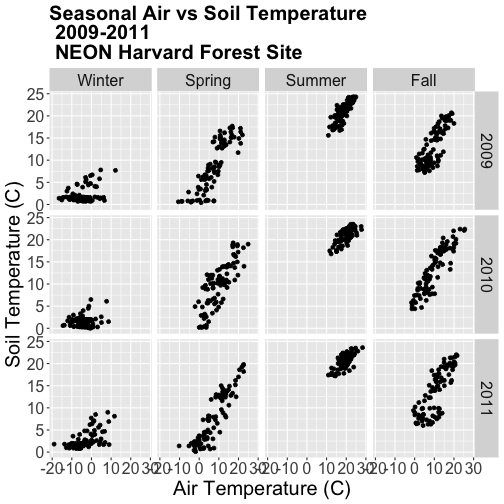
Work with Year-Month Data: base R and zoo Package
Some data will have month formatted in Year-Month
(e.g. met_monthly_HARV$date).
(Note: You will load this file in the Challenge below)
## [1] "2001-03" "2001-04" "2001-05" "2001-06" "2001-07" "2001-08"
For many analyses, we might want to summarize this data into a yearly total. Base R does NOT have a distinct year-month date class. Instead to work with a year-month field using base R, we need to convert to a Date class, which necessitates adding an associated day value. The syntax would be:
as.Date(paste(met_monthly_HARV$date,"-01", sep=""))
The syntax above creates a Date column from the met_montly_HARV$date column.
We then add the arbitrary date - the first ("-01"). The final bit of code
(sep="") designates the character string used to separate the month, day,
and year portions of the returned string (in our case nothing, as we have
included the hyphen with our inserted date value).
Alternatively, to work directly with a year-month data we could use the zoo
package and its included year-month date class - as.yearmon. With zoo the
syntax would be:
as.Date(as.yearmon(met_monthly_HARV$date))
Load the NEON-DS-Met-Time-Series/HARV/FisherTower-Met/hf001-04-monthly-m.csv
file and give it the name met_monthly_HARV. Then:
- Convert the date field into a date/time class using both base R and the
zoopackage. Name the new fieldsdate_baseandymon_zoorespectively. - Look at the format and check the class of both new date fields.
- Convert the
ymon_zoofield into a new Date class field (date_zoo) so it can be used in base R, ggplot, etc.
HINT: be sure to load the zoo package, if you have not already.
Do you prefer to use base R or zoo to convert these data to a date/time
class?
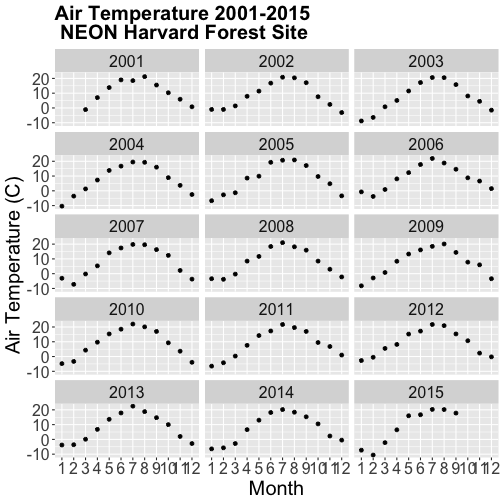
Get Lesson Code
Convert to Julian Day
Authors: Megan A. Jones, Marisa Guarinello, Courtney Soderberg, Leah A. Wasser
Last Updated: May 13, 2021
This tutorial defines Julian (year) day as most often used in an ecological context, explains why Julian days are useful for analysis and plotting, and teaches how to create a Julian day variable from a Date or Data/Time class variable.
Learning Objectives
After completing this tutorial, you will be able to:
- Define a Julian day (year day) as used in most ecological contexts.
- Convert a Date or Date/Time class variable to a Julian day variable.
Things You’ll Need To Complete This Tutorial
You will need the most current version of R and, preferably, RStudio loaded on your computer to complete this tutorial.
Install R Packages
-
lubridate:
install.packages("lubridate")
More on Packages in R – Adapted from Software Carpentry.
Download Data
NEON Teaching Data Subset: Meteorological Data for Harvard Forest
The data used in this lesson were collected at the National Ecological Observatory Network's Harvard Forest field site. These data are proxy data for what will be available for 30 years on the NEON data portal for the Harvard Forest and other field sites located across the United States.
Set Working Directory: This lesson assumes that you have set your working directory to the location of the downloaded and unzipped data subsets.
An overview of setting the working directory in R can be found here.
R Script & Challenge Code: NEON data lessons often contain challenges that reinforce learned skills. If available, the code for challenge solutions is found in the downloadable R script of the entire lesson, available in the footer of each lesson page.
Convert Between Time Formats - Julian Days
Julian days, as most often used in an ecological context, is a continuous count of the number of days beginning at Jan 1 each year. Thus each year will have up to 365 (non-leap year) or 366 (leap year) days.
Including a Julian day variable in your dataset can be very useful when comparing data across years, when plotting data, and when matching your data with other types of data that include Julian day.
Load the Data
Load this dataset that we will use to convert a date into a year day or Julian day.
Notice the date is read in as a character and must first be converted to a Date class.
# Load packages required for entire script
library(lubridate) #work with dates
# set working directory to ensure R can find the file we wish to import
wd <- "~/Git/data/"
# Load csv file of daily meteorological data from Harvard Forest
# Factors=FALSE so strings, series of letters/ words/ numerals, remain characters
harMet_DailyNoJD <- read.csv(
file=paste0(wd,"NEON-DS-Met-Time-Series/HARV/FisherTower-Met/hf001-06-daily-m-NoJD.csv"),
stringsAsFactors = FALSE
)
# what data class is the date column?
str(harMet_DailyNoJD$date)
## chr [1:5345] "2/11/01" "2/12/01" "2/13/01" "2/14/01" "2/15/01" ...
# convert "date" from chr to a Date class and specify current date format
harMet_DailyNoJD$date<- as.Date(harMet_DailyNoJD$date, "%m/%d/%y")
Convert with yday()
To quickly convert from from Date to Julian days, can we use yday(), a
function from the lubridate package.
# to learn more about it type
?yday
We want to create a new column in the existing data frame, titled julian, that
contains the Julian day data.
# convert with yday into a new column "julian"
harMet_DailyNoJD$julian <- yday(harMet_DailyNoJD$date)
# make sure it worked all the way through.
head(harMet_DailyNoJD$julian)
## [1] 42 43 44 45 46 47
tail(harMet_DailyNoJD$julian)
## [1] 268 269 270 271 272 273