Series
Introduction to Hyperspectral Remote Sensing Data in R
In this series, we go over the basics of working with NEON hyperspectral remote sensing data in R. We cover the principles of hyperspectral data, how to open hyperspectral data stored in HDF5 format in R and how to extract bands and create rasters in GeoTIFF format. Finally, we demonstrate how to extract a hyperspectral-spectral signature from a single pixel using R, and interactively compare the spectra from different land cover types.
Data used in this series are from the National Ecological Observatory Network (NEON) and are in HDF5 format.
Series Objectives
After completing the series you will:
- Understand the collection of hyperspectral remote sensing data and how they can be used
- Understand how HDF5 data can be used to store spatial data and the associated benefits of this format when working with large spatial data cubes
- Know how to extract metadata from HDF5 files
- Know how to plot a matrix as an image and a raster
- Understand how to extract and plot spectra from an HDF5 file
- Know how to work with groups and datasets within an HDF5 file
- Know how to export a spatially projected GeoTIFF
- Create a raster stack in R which can then be used to create RGB images from bands in a hyperspectral data cube
- Plot data spatially on a map
- Create basic vegetation indices, like NDVI, using raster-based calculations in R
Things You’ll Need To Complete This Series
Setup RStudio
To complete some of the tutorials in this series, you will need an updated version of R and, preferably, RStudio installed on your computer.
R is a programming language that specializes in statistical computing. It is a powerful tool for exploratory data analysis. To interact with R, we strongly recommend RStudio, an interactive development environment (IDE).
Download Data
Data can be downloaded from the NEON Data Portal, using the R neonUtilities::byTileAOP function in each tutorial. You will only need to download the data once, you will use the same data from the first lesson in all the subsequent lessons.
About Hyperspectral Remote Sensing Data
Authors: Leah A. Wasser
Last Updated: Sep 27, 2023
Learning Objectives
After completing this tutorial, you will be able to:
- Define hyperspectral remote sensing.
- Explain the fundamental principles of hyperspectral remote sensing data.
- Describe the key attributes that are required to effectively work with hyperspectral remote sensing data in tools like R or Python.
- Describe what a "band" is.
Mapping the Invisible
About Hyperspectral Remote Sensing Data
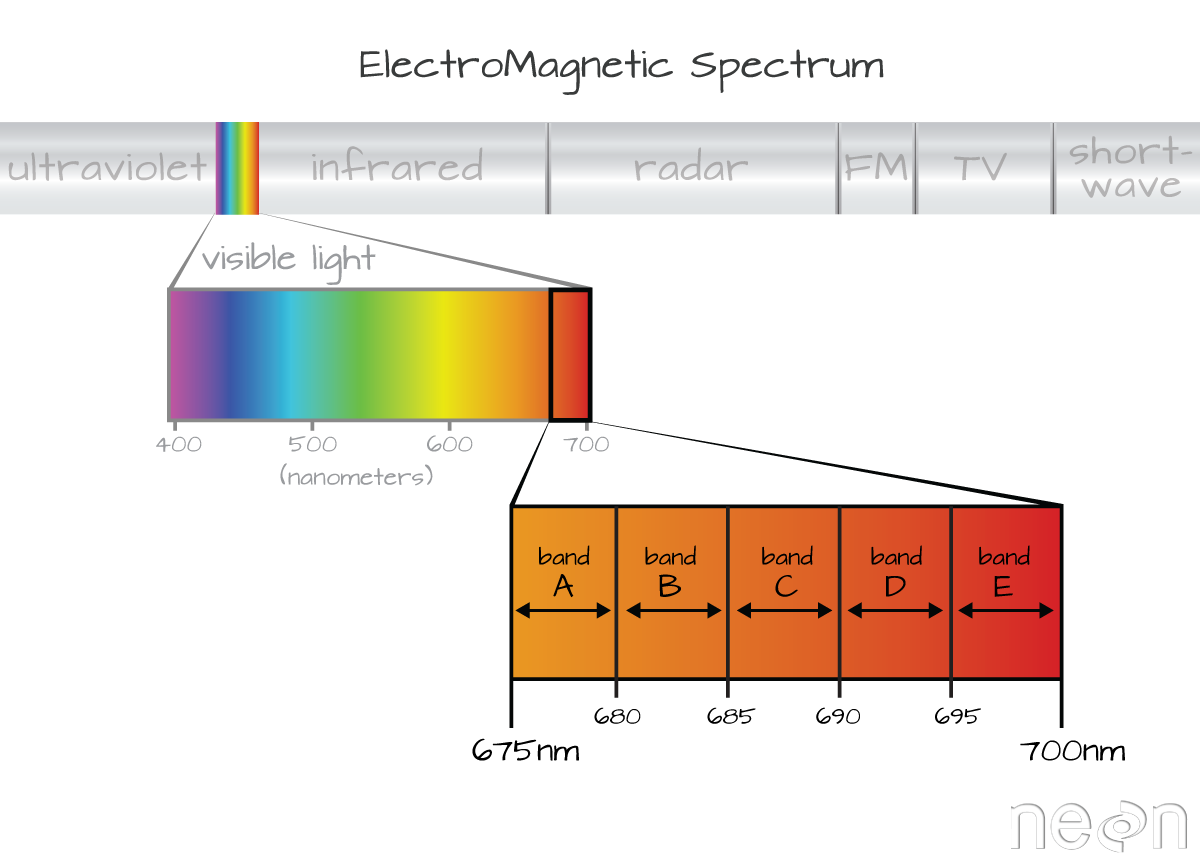
The electromagnetic spectrum is composed of thousands of bands representing different types of light energy. Imaging spectrometers (instruments that collect hyperspectral data) break the electromagnetic spectrum into groups of bands that support classification of objects by their spectral properties on the earth's surface. Hyperspectral data consists of many bands -- up to hundreds of bands -- that cover the electromagnetic spectrum.
The NEON imaging spectrometer collects data within the 380nm to 2510nm portions of the electromagnetic spectrum within bands that are approximately 5nm in width. This results in a hyperspectral data cube that contains approximately 426 bands - which means big, big data.
Key Metadata for Hyperspectral Data
Bands and Wavelengths
A band represents a group of wavelengths. For example, the wavelength values between 695nm and 700nm might be one band as captured by an imaging spectrometer. The imaging spectrometer collects reflected light energy in a pixel for light in that band. Often when you work with a multi or hyperspectral dataset, the band information is reported as the center wavelength value. This value represents the center point value of the wavelengths represented in that band. Thus in a band spanning 695-700 nm, the center would be 697.5).
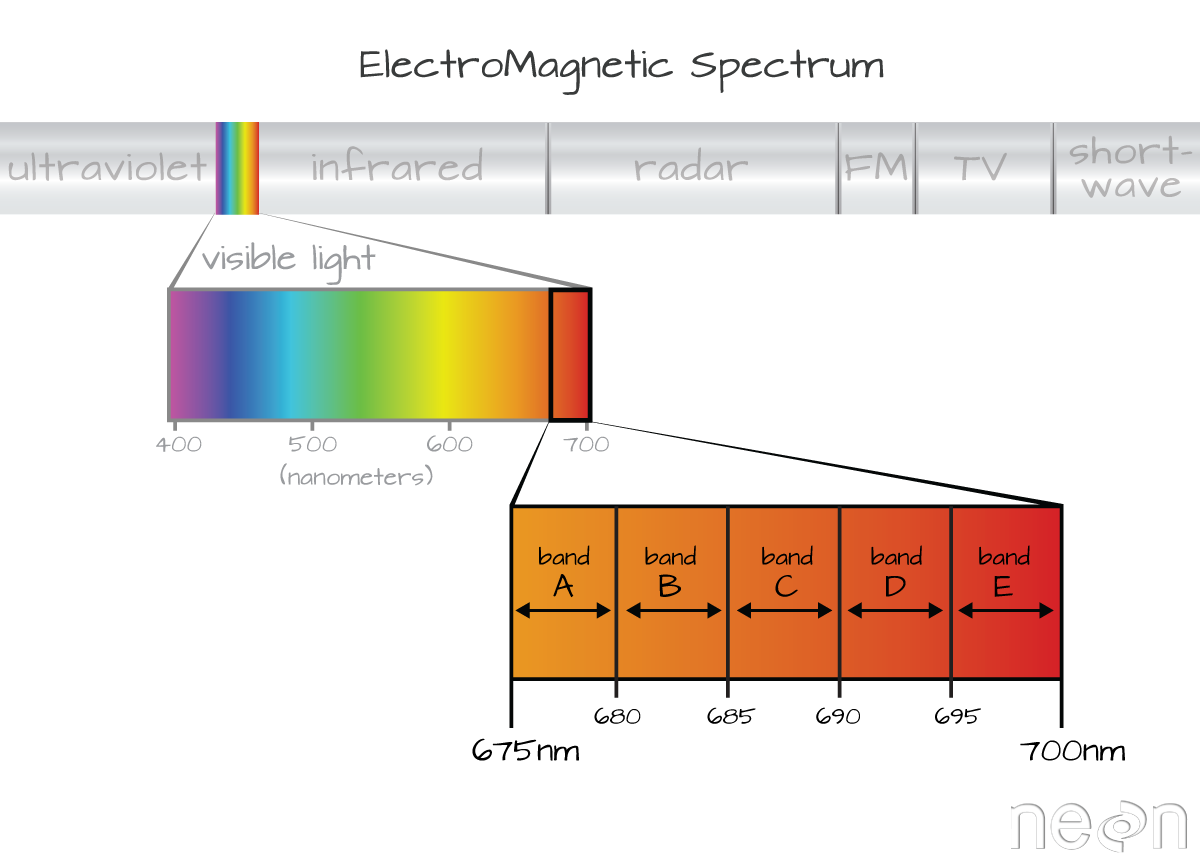
Spectral Resolution
The spectral resolution of a dataset that has more than one band, refers to the width of each band in the dataset. In the example above, a band was defined as spanning 695-700nm. The width or spatial resolution of the band is thus 5 nanometers. To see an example of this, check out the band widths for the Landsat sensors.
Full Width Half Max (FWHM)
The full width half max (FWHM) will also often be reported in a multi or hyperspectral dataset. This value represents the spread of the band around that center point.
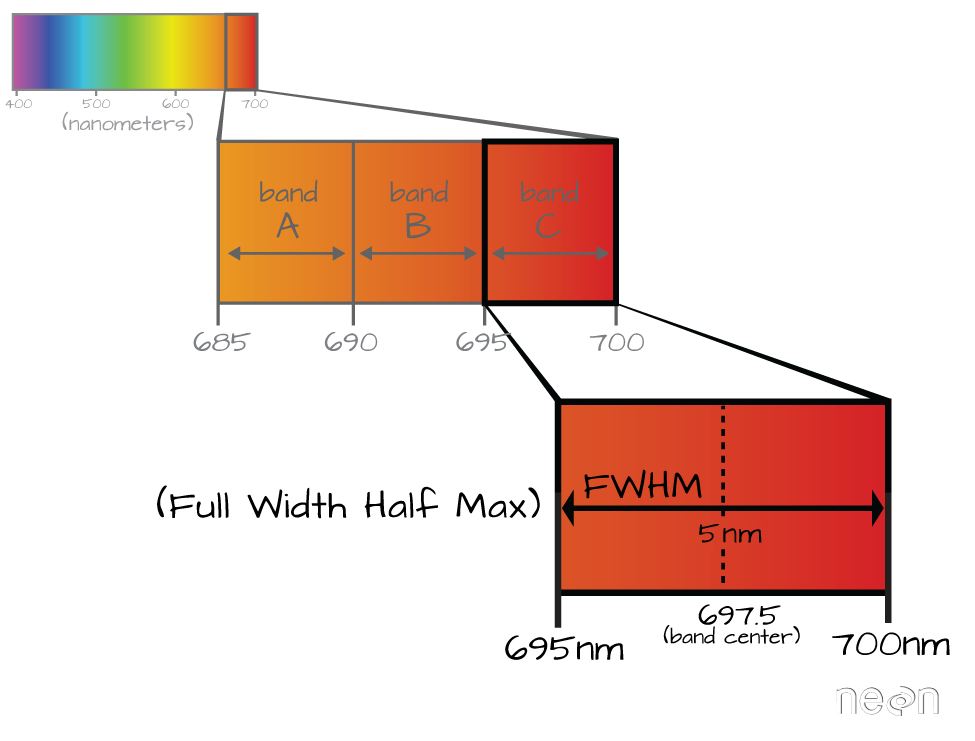
In the illustration above, the band that covers 695-700nm has a FWHM of 5 nm. While a general spectral resolution of the sensor is often provided, not all sensors create bands of uniform widths. For instance bands 1-9 of Landsat 8 are listed below (Courtesy of USGS)
| Band | Wavelength range (microns) | Spatial Resolution (m) | Spectral Width (microns) |
|---|---|---|---|
| Band 1 - Coastal aerosol | 0.43 - 0.45 | 30 | 0.02 |
| Band 2 - Blue | 0.45 - 0.51 | 30 | 0.06 |
| Band 3 - Green | 0.53 - 0.59 | 30 | 0.06 |
| Band 4 - Red | 0.64 - 0.67 | 30 | 0.03 |
| Band 5 - Near Infrared (NIR) | 0.85 - 0.88 | 30 | 0.03 |
| Band 6 - SWIR 1 | 1.57 - 1.65 | 30 | 0.08 |
| Band 7 - SWIR 2 | 2.11 - 2.29 | 30 | 0.18 |
| Band 8 - Panchromatic | 0.50 - 0.68 | 15 | 0.18 |
| Band 9 - Cirrus | 1.36 - 1.38 | 30 | 0.02 |
Data Product:
DP3.30006.001 | Spectrometer orthorectified surface directional reflectance - mosaicExplore Related Data Products
These data products are used in this tutorial:
Intro to Working with Hyperspectral Remote Sensing Data in HDF5 Format in R
Authors: Leah A. Wasser, Edmund Hart, Donal O'Leary
Last Updated: Jun 9, 2024
In this tutorial, we will show how to read and extract NEON reflectance data stored within an HDF5 file using R.
Learning Objectives
After completing this tutorial, you will be able to:
- Explain how HDF5 data can be used to store spatial data and the associated benefits of this format when working with large spatial data cubes.
- Extract metadata from HDF5 files.
- Slice or subset HDF5 data. You will extract one band of pixels.
- Plot a matrix as an image and a raster.
- Export a final GeoTIFF (spatially projected) that can be used both in further analysis and in common GIS tools like QGIS.
Things You’ll Need To Complete This Tutorial
To complete this tutorial you will need the most current version of R and, preferably, RStudio installed on your computer.
R Libraries to Install:
-
rhdf5:
install.packages("BiocManager"),BiocManager::install("rhdf5") -
terra:
install.packages("terra") -
neonUtilities:
install.packages("neonUtilities")
More on Packages in R - Adapted from Software Carpentry.
Data to Download
Data will be downloaded in the tutorial using the neonUtilities::byTileAOP function.
These hyperspectral remote sensing data provide information on the National Ecological Observatory Network's San Joaquin Experimental Range field site in March of 2021. The data were collected over the San Joaquin field site located in California (Domain 17).The entire dataset can be also be downloaded from the NEON Data Portal.
Set Working Directory: This lesson assumes that you have set your working directory to the location of the downloaded data.
An overview of setting the working directory in R can be found here.
R Script & Challenge Code: NEON data lessons often contain challenges to reinforce skills. If available, the code for challenge solutions is found in the downloadable R script of the entire lesson, available in the footer of each lesson page.
About Hyperspectral Remote Sensing Data
The electromagnetic spectrum is composed of thousands of bands representing different types of light energy. Imaging spectrometers (instruments that collect hyperspectral data) break the electromagnetic spectrum into groups of bands that support classification of objects by their spectral properties on the Earth's surface. Hyperspectral data consists of many bands - up to hundreds of bands - that span a portion of the electromagnetic spectrum, from the visible to the Short Wave Infrared (SWIR) regions.
The NEON imaging spectrometer (NIS) collects data within the 380 nm to 2510 nm portions of the electromagnetic spectrum within bands that are approximately 5 nm in width. This results in a hyperspectral data cube that contains approximately 426 bands - which means BIG DATA.
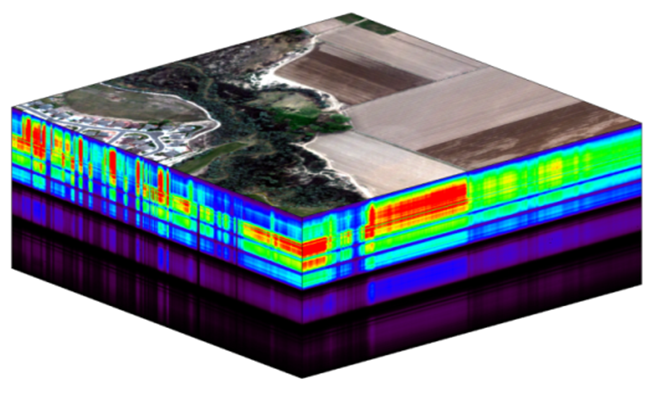
The HDF5 data model natively compresses data stored within it (makes it smaller) and supports data slicing (extracting only the portions of the data that you need to work with rather than reading the entire dataset into memory). These features make it ideal for working with large data cubes such as those generated by imaging spectrometers, in addition to supporting spatial data and associated metadata.
In this tutorial we will demonstrate how to read and extract spatial raster data stored within an HDF5 file using R.
Read HDF5 data into R
We will use the terra and rhdf5 packages to read in the HDF5 file that contains hyperspectral data for the
NEON San Joaquin (SJER) field site.
Let's start by calling the needed packages and reading in our NEON HDF5 file.
Please be sure that you have at least version 2.10 of rhdf5 installed. Use:
packageVersion("rhdf5") to check the package version.
Data Tip: To update all packages installed in R, use update.packages().
# Load `terra` and `rhdf5` packages to read NIS data into R
library(terra)
library(rhdf5)
library(neonUtilities)
Set the working directory to ensure R can find the file we are importing, and we know where the file is being saved. You can move the file that is downloaded afterward, but be sure to re-set the path to the file.
wd <- "~/data/" #This will depend on your local environment
setwd(wd)
We can use the neonUtilities function byTileAOP to download a single reflectance tile. You can run help(byTileAOP) to see more details on what the various inputs are. For this exercise, we'll specify the UTM Easting and Northing to be (257500, 4112500), which will download the tile with the lower left corner (257000,4112000). By default, the function will check the size total size of the download and ask you whether you wish to proceed (y/n). This file is ~672.7 MB, so make sure you have enough space on your local drive. You can set check.size=FALSE if you want to download without a prompt.
byTileAOP(dpID='DP3.30006.001',
site='SJER',
year='2021',
easting=257500,
northing=4112500,
check.size=TRUE, # set to FALSE if you don't want to enter y/n
savepath = wd)
This file will be downloaded into a nested subdirectory under the ~/data folder, inside a folder named DP3.30006.001 (the Data Product ID). The file should show up in this location: ~/data/DP3.30006.001/neon-aop-products/2021/FullSite/D17/2021_SJER_5/L3/Spectrometer/Reflectance/NEON_D17_SJER_DP3_257000_4112000_reflectance.h5.
Data Tip: To make sure you are pointing to the correct path, look in the ~/data folder and navigate to where the .h5 file is saved, or use the R command list.files(path=wd,pattern="\\.h5$",recursive=TRUE,full.names=TRUE) to display the full path of the .h5 file. Note, if you have any other .h5 files downloaded in this folder, it will display all of the hdf5 files.
# Define the h5 file name to be opened
h5_file <- paste0(wd,"DP3.30006.001/neon-aop-products/2021/FullSite/D17/2021_SJER_5/L3/Spectrometer/Reflectance/NEON_D17_SJER_DP3_257000_4112000_reflectance.h5")
You can use h5ls and/or View(h5ls(...)) to look at the contents of the hdf5 file, as follows:
# look at the HDF5 file structure
View(h5ls(h5_file,all=T))
When you look at the structure of the data, take note of the "map info" dataset, the Coordinate_System group, and the wavelength and Reflectance datasets. The Coordinate_System folder contains the spatial attributes of the data including its EPSG Code, which is easily converted to a Coordinate Reference System (CRS). The CRS documents how the data are physically located on the Earth. The wavelength dataset contains the wavelength values for each band in the data. The Reflectance dataset contains the image data that we will use for both data processing and visualization.
More Information on raster metadata:
-
Raster Data in R - The Basics - this tutorial explains more about how rasters work in R and their associated metadata.
-
About Hyperspectral Remote Sensing Data -this tutorial explains more about metadata and important concepts associated with multi-band (multi and hyperspectral) rasters.
Data Tip - HDF5 Structure: Note that the structure of individual HDF5 files may vary depending on who produced the data. In this case, the Wavelength and reflectance data within the file are both h5 datasets. However, the spatial information is contained within a group. Data downloaded from another organization (like NASA) may look different. This is why it's important to explore the data as a first step!
We can use the h5readAttributes() function to read and extract metadata from the HDF5 file. Let's start by learning about the wavelengths described within this file.
# get information about the wavelengths of this dataset
wavelengthInfo <- h5readAttributes(h5_file,"/SJER/Reflectance/Metadata/Spectral_Data/Wavelength")
wavelengthInfo
## $Description
## [1] "Central wavelength of the reflectance bands."
##
## $Units
## [1] "nanometers"
Next, we can use the h5read function to read the data contained within the HDF5 file. Let's read in the wavelengths of the band centers:
# read in the wavelength information from the HDF5 file
wavelengths <- h5read(h5_file,"/SJER/Reflectance/Metadata/Spectral_Data/Wavelength")
head(wavelengths)
## [1] 381.6035 386.6132 391.6229 396.6327 401.6424 406.6522
tail(wavelengths)
## [1] 2485.693 2490.703 2495.713 2500.722 2505.732 2510.742
Which wavelength is band 21 associated with?
(Hint: look at the wavelengths vector that we just imported and check out the data located at index 21 - wavelengths[21]).
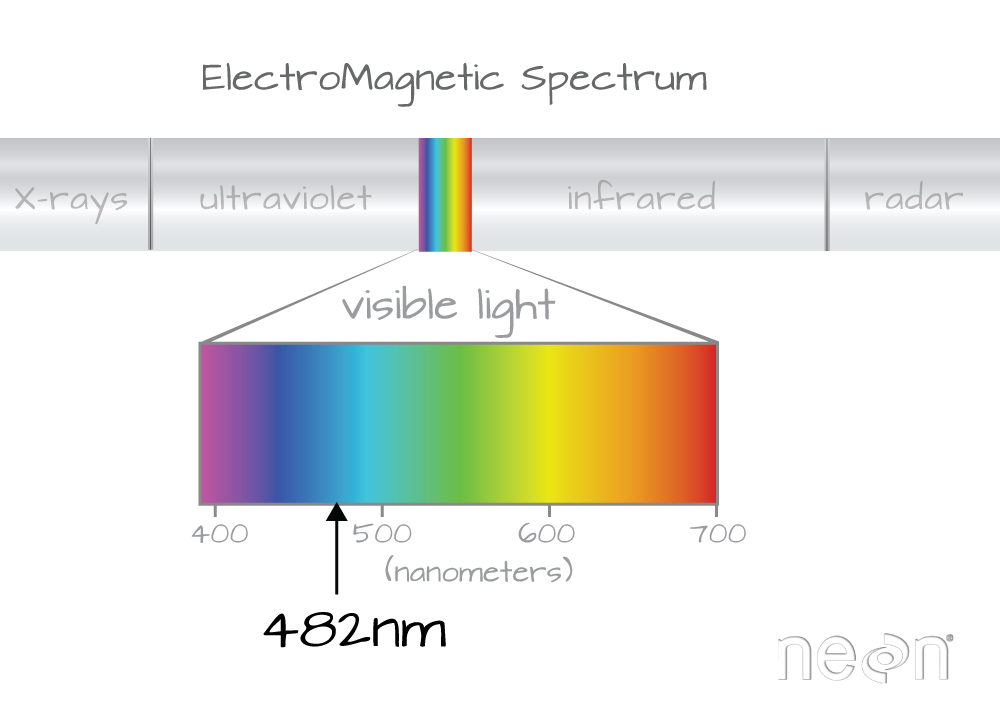
Band 21 has a associated wavelength center of 481.7982 nanometers (nm) which is in the blue portion (~380-500 nm) of the visible electromagnetic spectrum (~380-700 nm).
Bands and Wavelengths
A band represents a group of wavelengths. For example, the wavelength values between 695 nm and 700 nm might be one band captured by an imaging spectrometer. The imaging spectrometer collects reflected light energy in a pixel for light in that band. Often when you work with a multi- or hyperspectral dataset, the band information is reported as the center wavelength value. This value represents the mean value of the wavelengths represented in that band. Thus in a band spanning 695-700 nm, the center would be 697.5 nm). The full width half max (FWHM) will also be reported. This value can be thought of as the spread of the band around that center point. So, a band that covers 800-805 nm might have a FWHM of 5 nm and a wavelength value of 802.5 nm.
The HDF5 dataset that we are working with in this activity may contain more information than we need to work with. For example, we don't necessarily need to process all 426 bands available in a full NEON hyperspectral reflectance file - if we are interested in creating a product like NDVI which only uses bands in the Near InfraRed (NIR) and Red portions of the spectrum. Or we might only be interested in a spatial subset of the data - perhaps an area where we have collected corresponding ground data in the field.
The HDF5 format allows us to slice (or subset) the data - quickly extracting the subset that we need to process. Let's extract one of the green bands - band 34.
By the way - what is the center wavelength value associated with band 34?
Hint: wavelengths[34].
How do we know this band is a green band in the visible portion of the spectrum?
In order to effectively subset our data, let's first read the reflectance metadata stored as attributes in the "Reflectance_Data" dataset.
# First, we need to extract the reflectance metadata:
reflInfo <- h5readAttributes(h5_file, "/SJER/Reflectance/Reflectance_Data")
reflInfo
## $Cloud_conditions
## [1] "For cloud conditions information see Weather Quality Index dataset."
##
## $Cloud_type
## [1] "Cloud type may have been selected from multiple flight trajectories."
##
## $Data_Ignore_Value
## [1] -9999
##
## $Description
## [1] "Atmospherically corrected reflectance."
##
## $Dimension_Labels
## [1] "Line, Sample, Wavelength"
##
## $Dimensions
## [1] 1000 1000 426
##
## $Interleave
## [1] "BSQ"
##
## $Scale_Factor
## [1] 10000
##
## $Spatial_Extent_meters
## [1] 257000 258000 4112000 4113000
##
## $Spatial_Resolution_X_Y
## [1] 1 1
##
## $Units
## [1] "Unitless."
##
## $Units_Valid_range
## [1] 0 10000
# Next, we read the different dimensions
nRows <- reflInfo$Dimensions[1]
nCols <- reflInfo$Dimensions[2]
nBands <- reflInfo$Dimensions[3]
nRows
## [1] 1000
nCols
## [1] 1000
nBands
## [1] 426
The HDF5 read function reads data in the order: Bands, Cols, Rows. This is different from how R reads data. We'll adjust for this later.
# Extract or "slice" data for band 34 from the HDF5 file
b34 <- h5read(h5_file,"/SJER/Reflectance/Reflectance_Data",index=list(34,1:nCols,1:nRows))
# what type of object is b34?
class(b34)
## [1] "array"
A Note About Data Slicing in HDF5
Data slicing allows us to extract and work with subsets of the data rather than reading in the entire dataset into memory. In this example, we will extract and plot the green band without reading in all 426 bands. The ability to slice large datasets makes HDF5 ideal for working with big data.
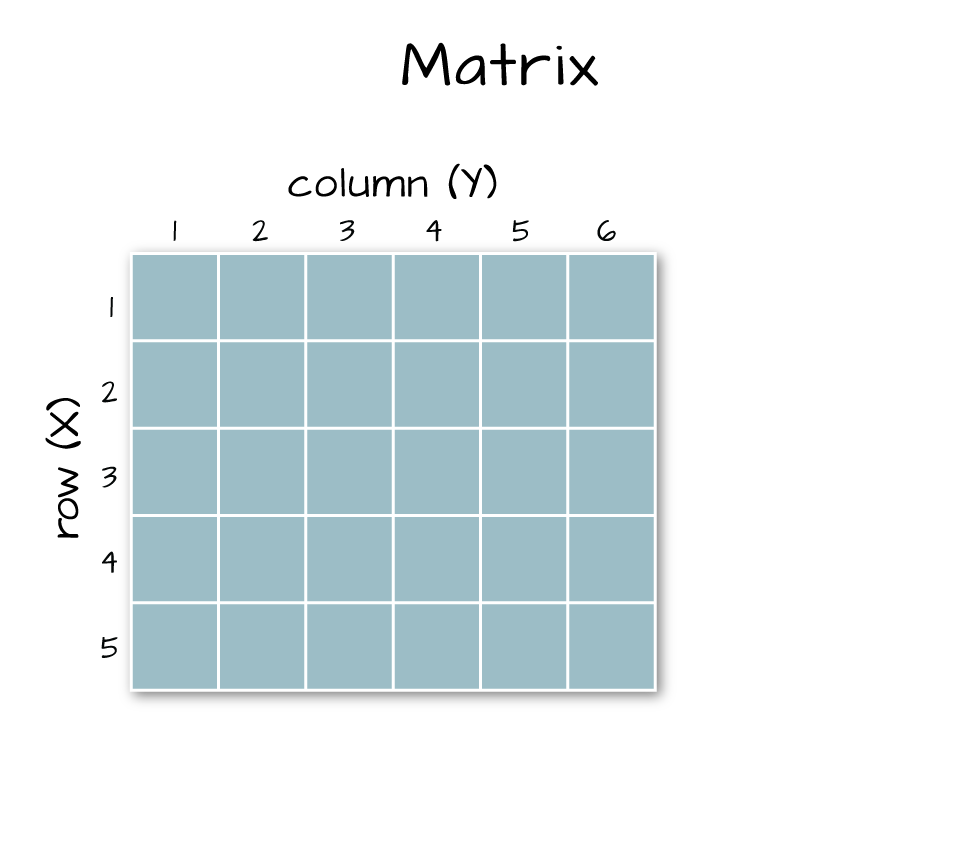
Next, let's convert our data from an array (more than 2 dimensions) to a matrix (just 2 dimensions). We need to have our data in a matrix format to plot it.
# convert from array to matrix by selecting only the first band
b34 <- b34[1,,]
# display the class of this re-defined variable
class(b34)
## [1] "matrix" "array"
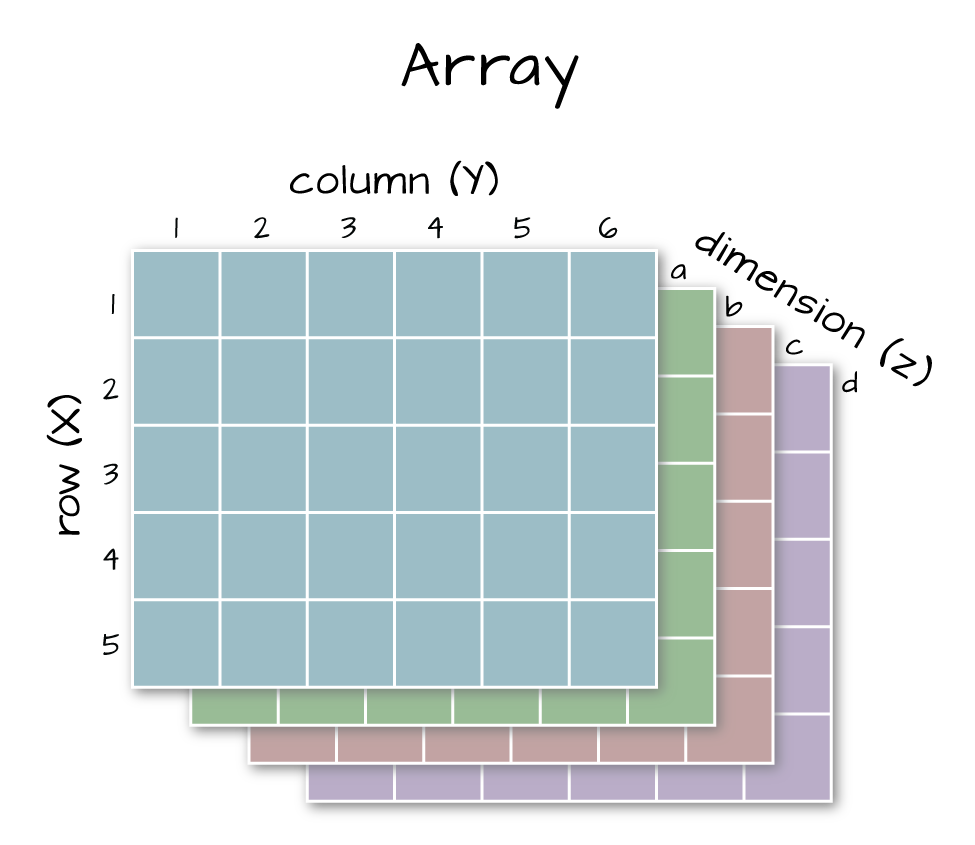
Arrays vs. Matrices
Arrays are matrices with more than 2 dimensions. When we say dimension, we are talking about the "z" associated with the data (imagine a series of tabs in a spreadsheet). Put the other way: matrices are arrays with only 2 dimensions. Arrays can have any number of dimensions one, two, ten or more.
Here is a matrix that is 4 x 3 in size (4 rows and 3 columns):
| Metric | species 1 | species 2 |
|---|---|---|
| total number | 23 | 45 |
| average weight | 14 | 5 |
| average length | 2.4 | 3.5 |
| average height | 32 | 12 |
Dimensions in Arrays
An array contains 1 or more dimensions in the "z" direction. For example, let's say that we collected the same set of species data for every day in a 30 day month. We might then have a matrix like the one above for each day for a total of 30 days making a 4 x 3 x 30 array (this dataset has more than 2 dimensions). More on R object types here (links to external site, DataCamp).
Next, let's look at the metadata for the reflectance data. When we do this, take note of 1) the scale factor and 2) the data ignore value. Then we can plot the band 34 data. Plotting spatial data as a visual "data check" is a good idea to make sure processing is being performed correctly and all is well with the image.
# look at the metadata for the reflectance dataset
h5readAttributes(h5_file,"/SJER/Reflectance/Reflectance_Data")
## $Cloud_conditions
## [1] "For cloud conditions information see Weather Quality Index dataset."
##
## $Cloud_type
## [1] "Cloud type may have been selected from multiple flight trajectories."
##
## $Data_Ignore_Value
## [1] -9999
##
## $Description
## [1] "Atmospherically corrected reflectance."
##
## $Dimension_Labels
## [1] "Line, Sample, Wavelength"
##
## $Dimensions
## [1] 1000 1000 426
##
## $Interleave
## [1] "BSQ"
##
## $Scale_Factor
## [1] 10000
##
## $Spatial_Extent_meters
## [1] 257000 258000 4112000 4113000
##
## $Spatial_Resolution_X_Y
## [1] 1 1
##
## $Units
## [1] "Unitless."
##
## $Units_Valid_range
## [1] 0 10000
# plot the image
image(b34)
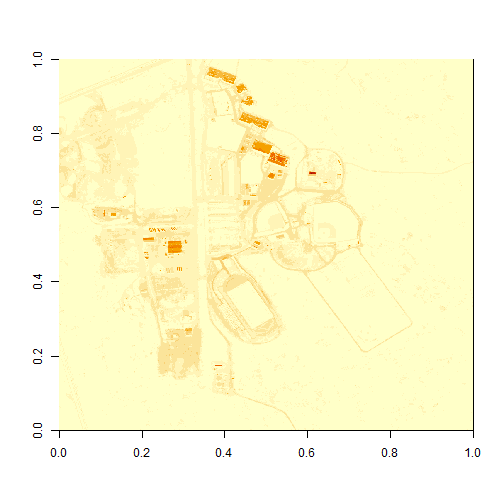
What do you notice about the first image? It's washed out and lacking any detail. What could be causing this? It got better when plotting the log of the values, but still not great.
# this is a little hard to visually interpret - what happens if we plot a log of the data?
image(log(b34))

Let's look at the distribution of reflectance values in our data to figure out what is going on.
# Plot range of reflectance values as a histogram to view range
# and distribution of values.
hist(b34,breaks=50,col="darkmagenta")
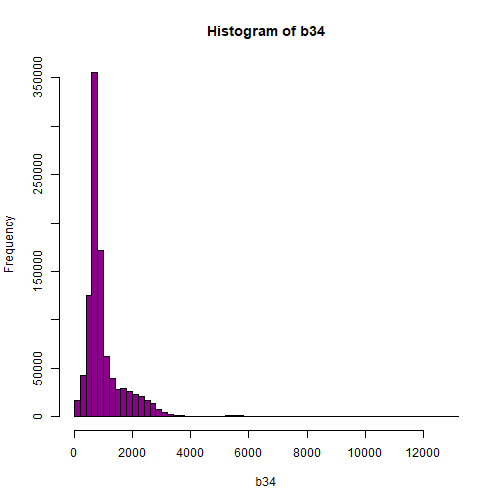
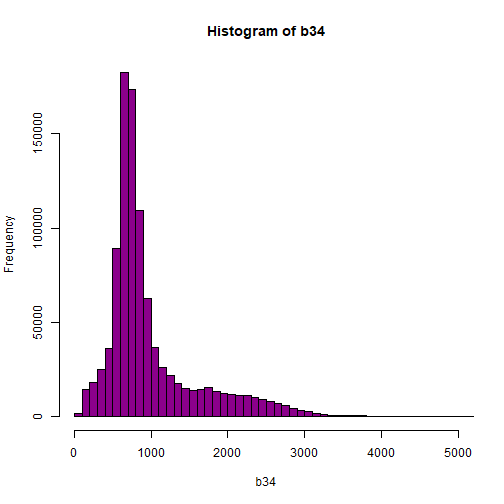
# View values between 0 and 5000
hist(b34,breaks=100,col="darkmagenta",xlim = c(0, 5000))
# View higher values
hist(b34, breaks=100,col="darkmagenta",xlim = c(5000, 15000),ylim = c(0, 750))
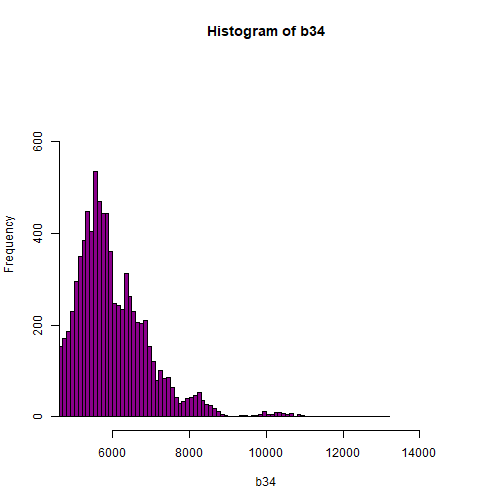
As you're examining the histograms above, keep in mind that reflectance values range between 0-1. The data scale factor in the metadata tells us to divide all reflectance values by 10,000. Thus, a value of 5,000 equates to a reflectance value of 0.50. Storing data as integers (without decimal places) compared to floating points (with decimal places) creates a smaller file. This type of scaling is commin in remote sensing datasets.
Notice in the data that there are some larger reflectance values (>5,000) that represent a smaller number of pixels. These pixels are skewing how the image renders.
Data Ignore Value
Image data in raster format will often contain a data ignore value and a scale factor. The data ignore value represents pixels where there are no data. Among other causes, no data values may be attributed to the sensor not collecting data in that area of the image or to processing results which yield null values.
Remember that the metadata for the Reflectance dataset designated -9999 as data ignore value. Thus, let's set all pixels with a value == -9999 to NA (no value). If we do this, R won't render these pixels.
# there is a no data value in our raster - let's define it
noDataValue <- as.numeric(reflInfo$Data_Ignore_Value)
noDataValue
## [1] -9999
# set all values equal to the no data value (-9999) to NA
b34[b34 == noDataValue] <- NA
# plot the image now
image(b34)
Reflectance Values and Image Stretch
Our image still looks dark because R is trying to render all reflectance values between 0 and 14999 as if they were distributed equally in the histogram. However we know they are not distributed equally. There are many more values between 0-5000 than there are values >5000.
Images contain a distribution of reflectance values. A typical image viewing program will render the values by distributing the entire range of reflectance values across a range of "shades" that the monitor can render - between 0 and 255. However, often the distribution of reflectance values is not linear. For example, in the case of our data, most of the reflectance values fall between 0 and 0.5. Yet there are a few values >0.8 that are heavily impacting the way the image is drawn on our monitor. Imaging processing programs like ENVI, QGIS and ArcGIS (and even Adobe Photoshop) allow you to adjust the stretch of the image. This is similar to adjusting the contrast and brightness in Photoshop.
The proper way to adjust our data would be to apply what's called an image stretch. We will learn how to stretch our image data later. For now, let's plot the values as the log function on the pixel reflectance values to factor out those larger values.
image(log(b34))

The log applied to our image increases the contrast making it look more like an image. However, look at the images below. The top one is an RGB image as the image should look. The bottom one is our log-adjusted image. Notice a difference?

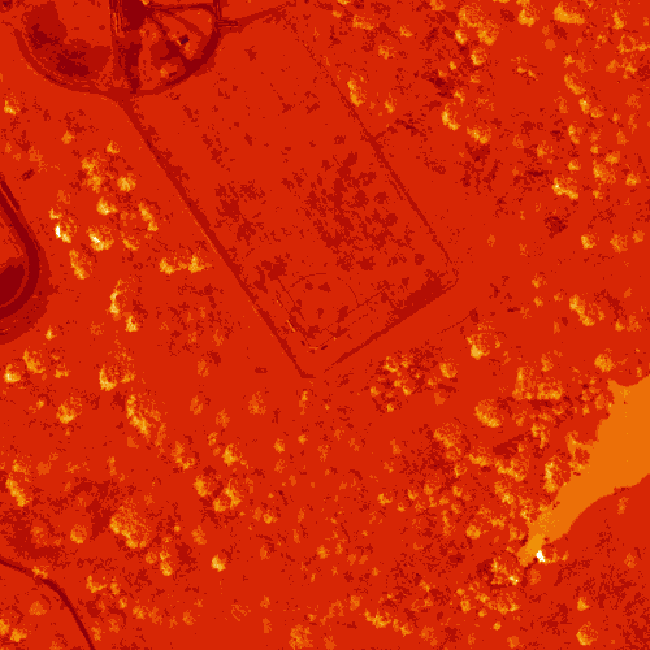
Transpose Image
Notice that there are three data dimensions for this file: Bands x Rows x Columns. However, when R reads in the dataset, it reads them as: Columns x Bands x Rows. The data are flipped. We can quickly transpose the data to correct for this using the t or transpose command in R.
The orientation is rotated in our log adjusted image. This is because R reads in matrices starting from the upper left hand corner. While most rasters read pixels starting from the lower left hand corner. In the next section, we will deal with this issue by creating a proper georeferenced (spatially located) raster in R. The raster format will read in pixels following the same methods as other GIS and imaging processing software like QGIS and ENVI do.
# We need to transpose x and y values in order for our
# final image to plot properly
b34 <- t(b34)
image(log(b34), main="Transposed Image")
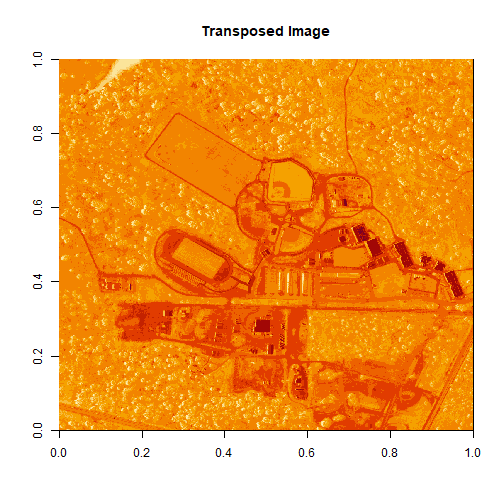
Create a Georeferenced Raster
Next, we will create a proper raster using the b34 matrix. The raster format will allow us to define and manage:
- Image stretch
- Coordinate reference system & spatial reference
- Resolution
- and other raster attributes...
It will also account for the orientation issue discussed above.
To create a raster in R, we need a few pieces of information, including:
- The coordinate reference system (CRS)
- The spatial extent of the image
Define Raster CRS
First, we need to define the Coordinate reference system (CRS) of the raster. To do that, we can first grab the EPSG code from the HDF5 attributes, and covert the EPSG to a CRS string. Then we can assign that CRS to the raster object.
# Extract the EPSG from the h5 dataset
h5EPSG <- h5read(h5_file, "/SJER/Reflectance/Metadata/Coordinate_System/EPSG Code")
# convert the EPSG code to a CRS string
h5CRS <- crs(paste0("+init=epsg:",h5EPSG))
# define final raster with projection info
# note that capitalization will throw errors on a MAC.
# if UTM is all caps it might cause an error!
b34r <- rast(b34,
crs=h5CRS)
# view the raster attributes
b34r
## class : SpatRaster
## dimensions : 1000, 1000, 1 (nrow, ncol, nlyr)
## resolution : 1, 1 (x, y)
## extent : 0, 1000, 0, 1000 (xmin, xmax, ymin, ymax)
## coord. ref. : WGS 84 / UTM zone 11N
## source(s) : memory
## name : lyr.1
## min value : 32
## max value : 13129
# let's have a look at our properly oriented raster. Take note of the
# coordinates on the x and y axis.
image(log(b34r),
xlab = "UTM Easting",
ylab = "UTM Northing",
main = "Properly Oriented Raster")
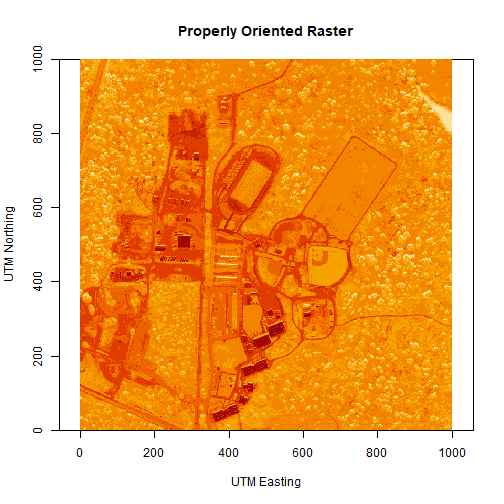
Next we define the extents of our raster. The extents will be used to calculate the raster's resolution. Fortunately, the spatial extent is provided in the HDF5 file "Reflectance_Data" group attributes that we saved before as reflInfo.
# Grab the UTM coordinates of the spatial extent
xMin <- reflInfo$Spatial_Extent_meters[1]
xMax <- reflInfo$Spatial_Extent_meters[2]
yMin <- reflInfo$Spatial_Extent_meters[3]
yMax <- reflInfo$Spatial_Extent_meters[4]
# define the extent (left, right, top, bottom)
rasExt <- ext(xMin,xMax,yMin,yMax)
rasExt
## SpatExtent : 257000, 258000, 4112000, 4113000 (xmin, xmax, ymin, ymax)
# assign the spatial extent to the raster
ext(b34r) <- rasExt
# look at raster attributes
b34r
## class : SpatRaster
## dimensions : 1000, 1000, 1 (nrow, ncol, nlyr)
## resolution : 1, 1 (x, y)
## extent : 257000, 258000, 4112000, 4113000 (xmin, xmax, ymin, ymax)
## coord. ref. : WGS 84 / UTM zone 11N
## source(s) : memory
## name : lyr.1
## min value : 32
## max value : 13129
Learn more about working with Raster data in R in the Data Carpentry workshop: Introduction to Geospatial Raster and Vector Data with R.
We can adjust the colors of our raster as well, if desired.
# let's change the colors of our raster and adjust the zlim
col <- terrain.colors(25)
image(b34r,
xlab = "UTM Easting",
ylab = "UTM Northing",
main= "Spatially Referenced Raster",
col=col,
zlim=c(0,3000))
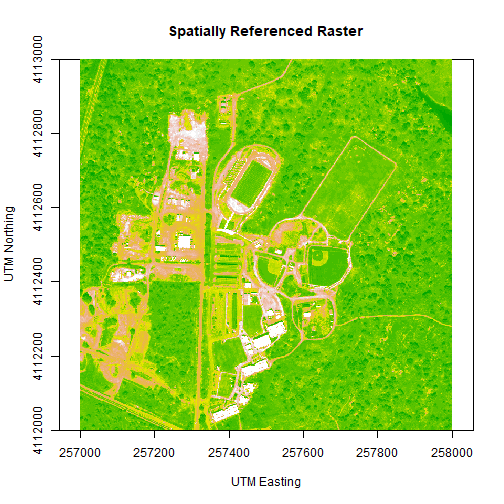
We've now created a raster from band 34 reflectance data. We can export the data as a raster, using the writeRaster command. Note that it's good practice to close the H5 connection before moving on!
# write out the raster as a geotiff
writeRaster(b34r,
file=paste0(wd,"band34.tif"),
overwrite=TRUE)
# close the H5 file
H5close()
Challenge: Work with Rasters
Try these three extensions on your own:
-
Create rasters using other bands in the dataset.
-
Vary the distribution of values in the image to mimic an image stretch. e.g.
b34[b34 > 6000 ] <- 6000 -
Use what you know to extract ALL of the reflectance values for ONE pixel rather than for an entire band. HINT: this will require you to pick an x and y value and then all values in the z dimension:
aPixel<- h5read(h5_file,"Reflectance",index=list(NULL,100,35)). Plot the spectra output.
Get Lesson Code
Creating a Raster Stack from Hyperspectral Imagery in HDF5 Format in R
Authors: Edmund Hart, Leah A. Wasser, Donal O'Leary
Last Updated: Feb 19, 2024
In this tutorial, we will learn how to create multi (3) band images from hyperspectral data. We will also learn how to perform some basic raster calculations (known as raster math in the GIS world).
Learning Objectives
After completing this activity, you will be able to:
- Extract a "slice" of data from a hyperspectral data cube.
- Create a raster "stack" in R which can be used to create RGB images from band combinations in a hyperspectral data cube.
- Plot data spatially on a map.
- Create basic vegetation indices like NDVI using raster-based calculations in R.
Things You’ll Need To Complete This Tutorial
To complete this tutorial you will need the most current version of R and, preferably, RStudio loaded on your computer.
R Libraries to Install:
-
rhdf5:
install.packages("BiocManager"),BiocManager::install("rhdf5") -
terra:
install.packages("terra") -
neonUtilities:
install.packages("neonUtilities")
More on Packages in R - Adapted from Software Carpentry.
Data
These hyperspectral remote sensing data provide information on the National Ecological Observatory Network's San Joaquin Experimental Range (SJER) field site in March of 2021. The data used in this lesson is the 1km by 1km mosaic tile named NEON_D17_SJER_DP3_257000_4112000_reflectance.h5. If you already completed the previous lesson in this tutorial series, you do not need to download this data again. The entire SJER reflectance dataset can be accessed from the NEON Data Portal.
Set Working Directory: This lesson assumes that you have set your working directory to the location of the downloaded data, as explained in the tutorial.
An overview of setting the working directory in R can be found here.
R Script & Challenge Code: NEON data lessons often contain challenges to reinforce skills. If available, the code for challenge solutions is found in the downloadable R script of the entire lesson, available in the footer of each lesson page.
Recommended Skills
For this tutorial you should be comfortable working with HDF5 files that contain hyperspectral data, including reading in reflectance values and associated metadata and attributes.
If you aren't familiar with these steps already, we highly recommend you work through the Introduction to Working with Hyperspectral Data in HDF5 Format in R tutorial before moving on to this tutorial.
About Hyperspectral Data
We often want to generate a 3 band image from multi or hyperspectral data. The most commonly recognized band combination is RGB which stands for Red, Green and Blue. RGB images are just like an image that your camera takes. But other band combinations can be useful too. For example, near infrared images highlight healthy vegetation, which makes it easy to classify or identify where vegetation is located on the ground.

Data Tip - Band Combinations: The Biodiversity Informatics group created a great interactive tool that lets you explore band combinations. Check it out. Learn more about band combinations using a great online tool from the American Museum of Natural History! (The tool requires Flash player.)
Create a Raster Stack in R
In the previous lesson, we exported a single band of the NEON Reflectance data from a HDF5 file. In this activity, we will create a full color image using 3 (red, green and blue - RGB) bands. We will follow many of the steps we followed in the Intro to Working with Hyperspectral Remote Sensing Data in HDF5 Format in R tutorial. These steps included loading required packages, downloading the data (optionally, you don't need to do this if you downloaded the data from the previous lesson), and reading in our file and viewing the hdf5 file structure.
First, let's load the required R packages, terra and rhdf5.
library(terra)
library(rhdf5)
library(neonUtilities)
Next set the working directory to ensure R can find the file we wish to import. Be sure to move the download into your working directory!
# set working directory (this will depend on your local environment)
wd <- "~/data/"
setwd(wd)
We can use the neonUtilities function byTileAOP to download a single reflectance tile. You can run help(byTileAOP) to see more details on what the various inputs are. For this exercise, we'll specify the UTM Easting and Northing to be (257500, 4112500), which will download the tile with the lower left corner (257000, 4112000).
byTileAOP(dpID = 'DP3.30006.001',
site = 'SJER',
year = '2021',
easting = 257500,
northing = 4112500,
savepath = wd)
This file will be downloaded into a nested subdirectory under the ~/data folder, inside a folder named DP3.30006.001 (the Data Product ID). The file should show up in this location: ~/data/DP3.30006.001/neon-aop-products/2021/FullSite/D17/2021_SJER_5/L3/Spectrometer/Reflectance/NEON_D17_SJER_DP3_257000_4112000_reflectance.h5.
Now we can read in the file. You can move this file to a different location, but make sure to change the path accordingly.
# Define the h5 file name to be opened
h5_file <- paste0(wd,"DP3.30006.001/neon-aop-products/2021/FullSite/D17/2021_SJER_5/L3/Spectrometer/Reflectance/NEON_D17_SJER_DP3_257000_4112000_reflectance.h5")
As in the last lesson, let's use View(h5ls) to take a look inside this hdf5 dataset:
View(h5ls(h5_file,all=T))
To spatially locate our raster data, we need a few key attributes:
- The coordinate reference system
- The spatial extent of the raster
We'll begin by grabbing these key attributes from the H5 file.
# define coordinate reference system from the EPSG code provided in the HDF5 file
h5EPSG <- h5read(h5_file,"/SJER/Reflectance/Metadata/Coordinate_System/EPSG Code" )
h5CRS <- crs(paste0("+init=epsg:",h5EPSG))
# get the Reflectance_Data attributes
reflInfo <- h5readAttributes(h5_file,"/SJER/Reflectance/Reflectance_Data" )
# Grab the UTM coordinates of the spatial extent
xMin <- reflInfo$Spatial_Extent_meters[1]
xMax <- reflInfo$Spatial_Extent_meters[2]
yMin <- reflInfo$Spatial_Extent_meters[3]
yMax <- reflInfo$Spatial_Extent_meters[4]
# define the extent (left, right, top, bottom)
rastExt <- ext(xMin,xMax,yMin,yMax)
# view the extent to make sure that it looks right
rastExt
## SpatExtent : 257000, 258000, 4112000, 4113000 (xmin, xmax, ymin, ymax)
# Finally, define the no data value for later
h5NoDataValue <- as.integer(reflInfo$Data_Ignore_Value)
cat('No Data Value:',h5NoDataValue)
## No Data Value: -9999
Next, we'll write a function that will perform the processing that we did step by step in the Intro to Working with Hyperspectral Remote Sensing Data in HDF5 Format in R. This will allow us to process multiple bands in bulk.
The function band2Rast slices a band of data from the HDF5 file, and extracts the reflectance array for that band. It then converts the data into a matrix, converts it to a raster, and finally returns a spatially corrected raster for the specified band.
The function requires the following variables:
- file: the hdf5 reflectance file
- band: the band number we wish to extract
- noDataValue: the noDataValue for the raster
-
extent: a terra spatial extent (
SpatExtent) object . - crs: the Coordinate Reference System for the raster
The function output is a spatially referenced, R terra object.
# file: the hdf5 file
# band: the band you want to process
# returns: a matrix containing the reflectance data for the specific band
band2Raster <- function(file, band, noDataValue, extent, CRS){
# first, read in the raster
out <- h5read(file,"/SJER/Reflectance/Reflectance_Data",index=list(band,NULL,NULL))
# Convert from array to matrix
out <- (out[1,,])
# transpose data to fix flipped row and column order
# depending upon how your data are formatted you might not have to perform this
# step.
out <- t(out)
# assign data ignore values to NA
# note, you might chose to assign values of 15000 to NA
out[out == noDataValue] <- NA
# turn the out object into a raster
outr <- rast(out,crs=CRS)
# assign the extents to the raster
ext(outr) <- extent
# return the terra raster object
return(outr)
}
Now that the function is created, we can create our list of rasters. The list specifies which bands (or dimensions in our hyperspectral dataset) we want to include in our raster stack. Let's start with a typical RGB (red, green, blue) combination. We will use bands 14, 9, and 4 (bands 58, 34, and 19 in a full NEON hyperspectral dataset).
Data Tip - wavelengths and bands: Remember that you can look at the wavelengths dataset in the HDF5 file to determine the center wavelength value for each band. Keep in mind that this data subset only includes every fourth band that is available in a full NEON hyperspectral dataset!
# create a list of the bands (R,G,B) we want to include in our stack
rgb <- list(58,34,19)
# lapply tells R to apply the function to each element in the list
rgb_rast <- lapply(rgb,FUN=band2Raster, file = h5_file,
noDataValue=h5NoDataValue,
ext=rastExt,
CRS=h5CRS)
Check out the properties or rgb_rast:
rgb_rast
## [[1]]
## class : SpatRaster
## dimensions : 1000, 1000, 1 (nrow, ncol, nlyr)
## resolution : 1, 1 (x, y)
## extent : 257000, 258000, 4112000, 4113000 (xmin, xmax, ymin, ymax)
## coord. ref. : WGS 84 / UTM zone 11N
## source(s) : memory
## name : lyr.1
## min value : 0
## max value : 14950
##
## [[2]]
## class : SpatRaster
## dimensions : 1000, 1000, 1 (nrow, ncol, nlyr)
## resolution : 1, 1 (x, y)
## extent : 257000, 258000, 4112000, 4113000 (xmin, xmax, ymin, ymax)
## coord. ref. : WGS 84 / UTM zone 11N
## source(s) : memory
## name : lyr.1
## min value : 32
## max value : 13129
##
## [[3]]
## class : SpatRaster
## dimensions : 1000, 1000, 1 (nrow, ncol, nlyr)
## resolution : 1, 1 (x, y)
## extent : 257000, 258000, 4112000, 4113000 (xmin, xmax, ymin, ymax)
## coord. ref. : WGS 84 / UTM zone 11N
## source(s) : memory
## name : lyr.1
## min value : 9
## max value : 11802
Note that it displays properties of 3 rasters. Finally, we can create a raster stack from our list of rasters as follows:
rgbStack <- rast(rgb_rast)
In the code chunk above, we used the lapply() function, which is a powerful, flexible way to apply a function (in this case, our band2Raster() function) multiple times. You can learn more about lapply() here.
NOTE: We are using the raster stack object in R to store several rasters that are of the same CRS and extent. This is a popular and convenient way to organize co-incident rasters.
Next, add the names of the bands to the raster so we can easily keep track of the bands in the list.
# Create a list of band names
bandNames <- paste("Band_",unlist(rgb),sep="")
# set the rasterStack's names equal to the list of bandNames created above
names(rgbStack) <- bandNames
# check properties of the raster list - note the band names
rgbStack
## class : SpatRaster
## dimensions : 1000, 1000, 3 (nrow, ncol, nlyr)
## resolution : 1, 1 (x, y)
## extent : 257000, 258000, 4112000, 4113000 (xmin, xmax, ymin, ymax)
## coord. ref. : WGS 84 / UTM zone 11N
## source(s) : memory
## names : Band_58, Band_34, Band_19
## min values : 0, 32, 9
## max values : 14950, 13129, 11802
# scale the data as specified in the reflInfo$Scale Factor
rgbStack <- rgbStack/as.integer(reflInfo$Scale_Factor)
# plot one raster in the stack to make sure things look OK.
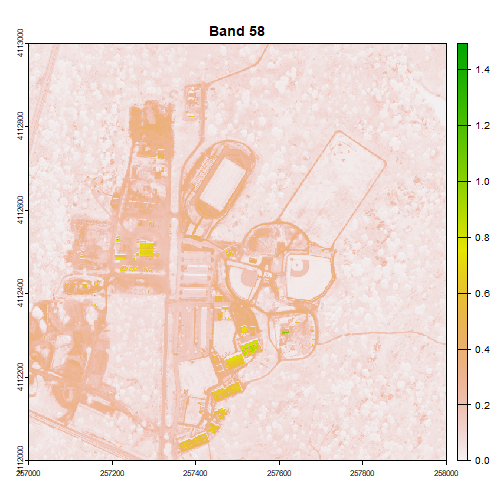
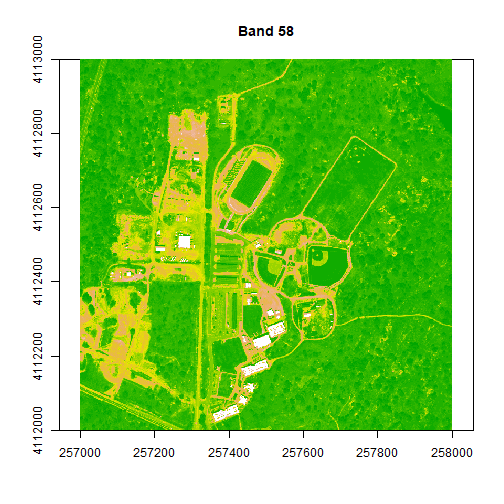
plot(rgbStack$Band_58, main="Band 58")
We can play with the color ramps too if we want:
# change the colors of our raster
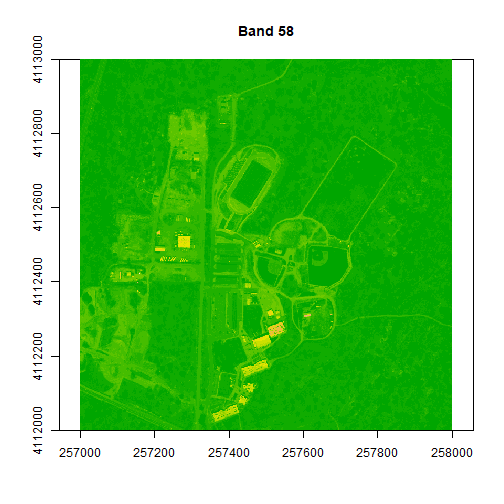
colors1 <- terrain.colors(25)
image(rgbStack$Band_58, main="Band 58", col=colors1)
# adjust the zlims or the stretch of the image
image(rgbStack$Band_58, main="Band 58", col=colors1, zlim = c(0,.5))
# try a different color palette
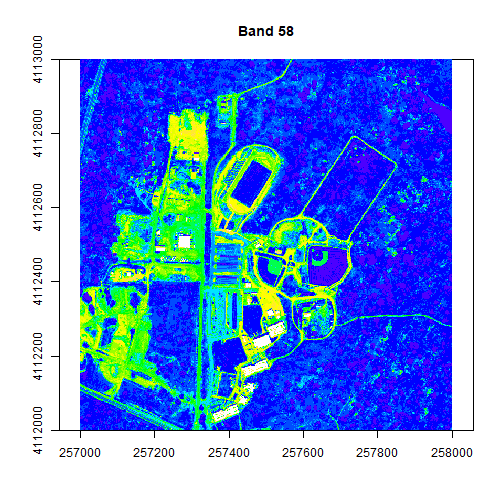
colors2 <- topo.colors(15, alpha = 1)
image(rgbStack$Band_58, main="Band 58", col=colors2, zlim=c(0,.5))
The plotRGB function allows you to combine three bands to create an true-color image.
# create a 3 band RGB image
plotRGB(rgbStack,
r=1,g=2,b=3,
stretch = "lin")

A note about image stretching: Notice that we use the argument stretch="lin" in this plotting function, which automatically stretches the brightness values for us to produce a natural-looking image.
Once you've created your raster, you can export it as a GeoTIFF using writeRaster. You can bring this GeoTIFF into any GIS software, such as QGIS or ArcGIS.
# Write out final raster
# Note: if you set overwrite to TRUE, then you will overwrite (and lose) any older version of the tif file!
writeRaster(rgbStack, file=paste0(wd,"NEON_hyperspectral_tutorial_example_RGB_image.tif"), overwrite=TRUE)
Data Tip - False color and near infrared images: Use the band combinations listed at the top of this page to modify the raster list. What type of image do you get when you change the band values?
Challenge: Other band combinations
Use different band combinations to create other "RGB" images. Suggested band combinations are below for use with the full NEON hyperspectral reflectance datasets (for this example dataset, divide the band number by 4 and round to the nearest whole number):
- Color Infrared/False Color: rgb (90,34,19)
- SWIR, NIR, Red Band: rgb (152,90,58)
- False Color: rgb (363,246,55)
Raster Math - Creating NDVI and other Vegetation Indices in R
In this last part, we will calculate some vegetation indices using raster math in R! We will start by creating NDVI or Normalized Difference Vegetation Index.
About NDVI
NDVI is a ratio between the near infrared (NIR) portion of the electromagnetic spectrum and the red portion of the spectrum.
$$ NDVI = \frac{NIR-RED}{NIR+RED} $$
Please keep in mind that there are different ways to aggregate bands when using hyperspectral data. This example is using individual bands to perform the NDVI calculation. Using individual bands is not necessarily the best way to calculate NDVI from hyperspectral data.
# Calculate NDVI
# select bands to use in calculation (red, NIR)
ndviBands <- c(58,90)
# create raster list and then a stack using those two bands
ndviRast <- lapply(ndviBands,FUN=band2Raster, file = h5_file,
noDataValue=h5NoDataValue,
ext=rastExt, CRS=h5CRS)
ndviStack <- rast(ndviRast)
# make the names pretty
bandNDVINames <- paste("Band_",unlist(ndviBands),sep="")
names(ndviStack) <- bandNDVINames
# view the properties of the new raster stack
ndviStack
## class : SpatRaster
## dimensions : 1000, 1000, 2 (nrow, ncol, nlyr)
## resolution : 1, 1 (x, y)
## extent : 257000, 258000, 4112000, 4113000 (xmin, xmax, ymin, ymax)
## coord. ref. : WGS 84 / UTM zone 11N
## source(s) : memory
## names : Band_58, Band_90
## min values : 0, 11
## max values : 14950, 14887
#calculate NDVI
NDVI <- function(x) {
(x[,2]-x[,1])/(x[,2]+x[,1])
}
ndviCalc <- app(ndviStack,NDVI)
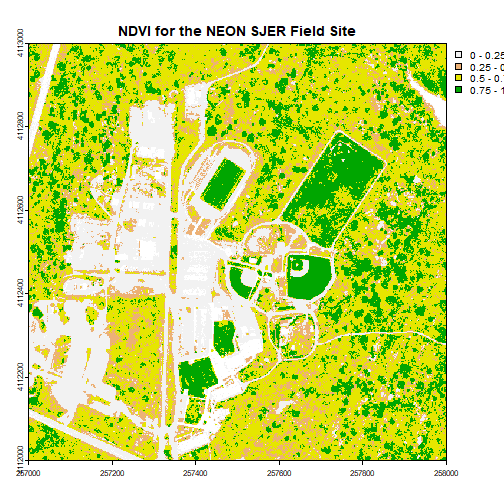
plot(ndviCalc, main="NDVI for the NEON SJER Field Site")

# Now, play with breaks and colors to create a meaningful map
# add a color map with 4 colors
myCol <- rev(terrain.colors(4)) # use the 'rev()' function to put green as the highest NDVI value
# add breaks to the colormap, including lowest and highest values (4 breaks = 3 segments)
brk <- c(0, .25, .5, .75, 1)
# plot the image using breaks
plot(ndviCalc, main="NDVI for the NEON SJER Field Site", col=myCol, breaks=brk)
Challenge: Work with Indices
Try the following on your own:
- Calculate the Normalized Difference Nitrogen Index (NDNI) using the following equation:
$$ NDNI = \frac{log(\frac{1}{p_{1510}}) - log(\frac{1}{p_{1680}})}{log(\frac{1}{p_{1510}}) + log(\frac{1}{p_{1680}})} $$
-
Calculate the Enhanced Vegetation Index (EVI). Hint: Look up the formula, and apply the appropriate NEON bands. Hint: You can look at satellite datasets, such as USGS Landsat EVI.
-
Explore the bands in the hyperspectral data. What happens if you average reflectance values across multiple Red and NIR bands and then calculate NDVI?
Get Lesson Code
Plot Spectral Signatures Derived from Hyperspectral Remote Sensing Data in HDF5 Format in R
Authors: Leah A. Wasser, Donal O'Leary
Last Updated: Feb 5, 2024
Learning Objectives
After completing this tutorial, you will be able to:
- Extract and plot a single spectral signature from an HDF5 file.
- Work with groups and datasets within an HDF5 file.
Things You’ll Need To Complete This Tutorial
To complete this tutorial you will need the most current version of R and, preferably, RStudio loaded on your computer.
R Libraries to Install:
-
rhdf5:
install.packages("BiocManager"),BiocManager::install("rhdf5") -
plyr:
install.packages('plyr') -
ggplot2:
install.packages('ggplot2') -
neonUtilities:
install.packages('neonUtilities')
More on Packages in R - Adapted from Software Carpentry.
Data
These hyperspectral remote sensing data provide information on the National Ecological Observatory Network's San Joaquin Experimental Range (SJER) field site in March of 2021. The data used in this lesson is the 1km by 1km mosaic tile named NEON_D17_SJER_DP3_257000_4112000_reflectance.h5. If you already completed the previous lesson in this tutorial series, you do not need to download this data again. The entire SJER reflectance dataset can be accessed from the NEON Data Portal.
Set Working Directory: This lesson assumes that you have set your working directory to the location of the downloaded and unzipped data subsets.
An overview of setting the working directory in R can be found here.
Recommended Skills
For this tutorial, you should be comfortable reading data from a HDF5 file and have a general familiarity with hyperspectral data. If you aren't familiar with these steps already, we highly recommend you work through the Introduction to Working with Hyperspectral Data in HDF5 Format in R tutorial before moving on to this tutorial.
Everything on our planet reflects electromagnetic radiation from the Sun, and different types of land cover often have dramatically different reflectance properties across the spectrum. One of the most powerful aspects of the NEON Imaging Spectrometer (NIS, or hyperspectral sensor) is that it can accurately measure these reflectance properties at a very high spectral resolution. When you plot the reflectance values across the observed spectrum, you will see that different land cover types (vegetation, pavement, bare soils, etc.) have distinct patterns in their reflectance values, a property that we call the 'spectral signature' of a particular land cover class.
In this tutorial, we will extract the reflectance values for all bands of a single pixel to plot a spectral signature for that pixel. In order to do this, we need to pair the reflectance values for that pixel with the wavelength values of the bands that are represented in those measurements. We will also need to adjust the reflectance values by the scaling factor that is saved as an 'attribute' in the HDF5 file. First, let's start by defining the working directory and reading in the example dataset.
# Call required packages
library(rhdf5)
library(plyr)
library(ggplot2)
library(neonUtilities)
wd <- "~/data/" #This will depend on your local environment
setwd(wd)
If you haven't already downloaded the hyperspectral data tile (in one of the previous tutorials in this series), you can use the neonUtilities function byTileAOP to download a single reflectance tile. You can run help(byTileAOP) to see more details on what the various inputs are. For this exercise, we'll specify the UTM Easting and Northing to be (257500, 4112500), which will download the tile with the lower left corner (257000, 4112000).
byTileAOP(dpID = 'DP3.30006.001',
site = 'SJER',
year = '2021',
easting = 257500,
northing = 4112500,
savepath = wd)
This file will be downloaded into a nested subdirectory under the ~/data folder (your working directory), inside a folder named DP3.30006.001 (the Data Product ID). The file should show up in this location: ~/data/DP3.30006.001/neon-aop-products/2021/FullSite/D17/2021_SJER_5/L3/Spectrometer/Reflectance/NEON_D17_SJER_DP3_257000_4112000_reflectance.h5.
Now we can read in the file and look at the contents using h5ls. You can move this file to a different location, but make sure to change the path accordingly.
# define the h5 file name (specify the full path)
h5_file <- paste0(wd,"DP3.30006.001/neon-aop-products/2021/FullSite/D17/2021_SJER_5/L3/Spectrometer/Reflectance/NEON_D17_SJER_DP3_257000_4112000_reflectance.h5")
# look at the HDF5 file structure
h5ls(h5_file) #optionally specify all=True if you want to see all of the information
## group name otype dclass dim
## 0 / SJER H5I_GROUP
## 1 /SJER Reflectance H5I_GROUP
## 2 /SJER/Reflectance Metadata H5I_GROUP
## 3 /SJER/Reflectance/Metadata Ancillary_Imagery H5I_GROUP
## 4 /SJER/Reflectance/Metadata/Ancillary_Imagery Aerosol_Optical_Depth H5I_DATASET INTEGER 1000 x 1000
## 5 /SJER/Reflectance/Metadata/Ancillary_Imagery Aspect H5I_DATASET FLOAT 1000 x 1000
## 6 /SJER/Reflectance/Metadata/Ancillary_Imagery Cast_Shadow H5I_DATASET INTEGER 1000 x 1000
## 7 /SJER/Reflectance/Metadata/Ancillary_Imagery Dark_Dense_Vegetation_Classification H5I_DATASET INTEGER 1000 x 1000
## 8 /SJER/Reflectance/Metadata/Ancillary_Imagery Data_Selection_Index H5I_DATASET INTEGER 1000 x 1000
## 9 /SJER/Reflectance/Metadata/Ancillary_Imagery Haze_Cloud_Water_Map H5I_DATASET INTEGER 1000 x 1000
## 10 /SJER/Reflectance/Metadata/Ancillary_Imagery Illumination_Factor H5I_DATASET INTEGER 1000 x 1000
## 11 /SJER/Reflectance/Metadata/Ancillary_Imagery Path_Length H5I_DATASET FLOAT 1000 x 1000
## 12 /SJER/Reflectance/Metadata/Ancillary_Imagery Sky_View_Factor H5I_DATASET INTEGER 1000 x 1000
## 13 /SJER/Reflectance/Metadata/Ancillary_Imagery Slope H5I_DATASET FLOAT 1000 x 1000
## 14 /SJER/Reflectance/Metadata/Ancillary_Imagery Smooth_Surface_Elevation H5I_DATASET FLOAT 1000 x 1000
## 15 /SJER/Reflectance/Metadata/Ancillary_Imagery Visibility_Index_Map H5I_DATASET INTEGER 1000 x 1000
## 16 /SJER/Reflectance/Metadata/Ancillary_Imagery Water_Vapor_Column H5I_DATASET FLOAT 1000 x 1000
## 17 /SJER/Reflectance/Metadata/Ancillary_Imagery Weather_Quality_Indicator H5I_DATASET INTEGER 3 x 1000 x 1000
## 18 /SJER/Reflectance/Metadata Coordinate_System H5I_GROUP
## 19 /SJER/Reflectance/Metadata/Coordinate_System Coordinate_System_String H5I_DATASET STRING ( 0 )
## 20 /SJER/Reflectance/Metadata/Coordinate_System EPSG Code H5I_DATASET STRING ( 0 )
## 21 /SJER/Reflectance/Metadata/Coordinate_System Map_Info H5I_DATASET STRING ( 0 )
## 22 /SJER/Reflectance/Metadata/Coordinate_System Proj4 H5I_DATASET STRING ( 0 )
## 23 /SJER/Reflectance/Metadata Flight_Trajectory H5I_GROUP
## 24 /SJER/Reflectance/Metadata Logs H5I_GROUP
## 25 /SJER/Reflectance/Metadata/Logs 195724 H5I_GROUP
## 26 /SJER/Reflectance/Metadata/Logs/195724 ATCOR_Input_file H5I_DATASET STRING ( 0 )
## 27 /SJER/Reflectance/Metadata/Logs/195724 ATCOR_Processing_Log H5I_DATASET STRING ( 0 )
## 28 /SJER/Reflectance/Metadata/Logs/195724 Shadow_Processing_Log H5I_DATASET STRING ( 0 )
## 29 /SJER/Reflectance/Metadata/Logs/195724 Skyview_Processing_Log H5I_DATASET STRING ( 0 )
## 30 /SJER/Reflectance/Metadata/Logs/195724 Solar_Azimuth_Angle H5I_DATASET FLOAT ( 0 )
## 31 /SJER/Reflectance/Metadata/Logs/195724 Solar_Zenith_Angle H5I_DATASET FLOAT ( 0 )
## 32 /SJER/Reflectance/Metadata/Logs 200251 H5I_GROUP
## 33 /SJER/Reflectance/Metadata/Logs/200251 ATCOR_Input_file H5I_DATASET STRING ( 0 )
## 34 /SJER/Reflectance/Metadata/Logs/200251 ATCOR_Processing_Log H5I_DATASET STRING ( 0 )
## 35 /SJER/Reflectance/Metadata/Logs/200251 Shadow_Processing_Log H5I_DATASET STRING ( 0 )
## 36 /SJER/Reflectance/Metadata/Logs/200251 Skyview_Processing_Log H5I_DATASET STRING ( 0 )
## 37 /SJER/Reflectance/Metadata/Logs/200251 Solar_Azimuth_Angle H5I_DATASET FLOAT ( 0 )
## 38 /SJER/Reflectance/Metadata/Logs/200251 Solar_Zenith_Angle H5I_DATASET FLOAT ( 0 )
## 39 /SJER/Reflectance/Metadata/Logs 200812 H5I_GROUP
## 40 /SJER/Reflectance/Metadata/Logs/200812 ATCOR_Input_file H5I_DATASET STRING ( 0 )
## 41 /SJER/Reflectance/Metadata/Logs/200812 ATCOR_Processing_Log H5I_DATASET STRING ( 0 )
## 42 /SJER/Reflectance/Metadata/Logs/200812 Shadow_Processing_Log H5I_DATASET STRING ( 0 )
## 43 /SJER/Reflectance/Metadata/Logs/200812 Skyview_Processing_Log H5I_DATASET STRING ( 0 )
## 44 /SJER/Reflectance/Metadata/Logs/200812 Solar_Azimuth_Angle H5I_DATASET FLOAT ( 0 )
## 45 /SJER/Reflectance/Metadata/Logs/200812 Solar_Zenith_Angle H5I_DATASET FLOAT ( 0 )
## 46 /SJER/Reflectance/Metadata/Logs 201441 H5I_GROUP
## 47 /SJER/Reflectance/Metadata/Logs/201441 ATCOR_Input_file H5I_DATASET STRING ( 0 )
## 48 /SJER/Reflectance/Metadata/Logs/201441 ATCOR_Processing_Log H5I_DATASET STRING ( 0 )
## 49 /SJER/Reflectance/Metadata/Logs/201441 Shadow_Processing_Log H5I_DATASET STRING ( 0 )
## 50 /SJER/Reflectance/Metadata/Logs/201441 Skyview_Processing_Log H5I_DATASET STRING ( 0 )
## 51 /SJER/Reflectance/Metadata/Logs/201441 Solar_Azimuth_Angle H5I_DATASET FLOAT ( 0 )
## 52 /SJER/Reflectance/Metadata/Logs/201441 Solar_Zenith_Angle H5I_DATASET FLOAT ( 0 )
## 53 /SJER/Reflectance/Metadata Spectral_Data H5I_GROUP
## 54 /SJER/Reflectance/Metadata/Spectral_Data FWHM H5I_DATASET FLOAT 426
## 55 /SJER/Reflectance/Metadata/Spectral_Data Wavelength H5I_DATASET FLOAT 426
## 56 /SJER/Reflectance/Metadata to-sensor_azimuth_angle H5I_DATASET FLOAT 1000 x 1000
## 57 /SJER/Reflectance/Metadata to-sensor_zenith_angle H5I_DATASET FLOAT 1000 x 1000
## 58 /SJER/Reflectance Reflectance_Data H5I_DATASET INTEGER 426 x 1000 x 1000
Read Wavelength Values
Next, let's read in the wavelength center associated with each band in the HDF5 file. We will later match these with the reflectance values and show both in our final spectral signature plot.
# read in the wavelength information from the HDF5 file
wavelengths <- h5read(h5_file,"/SJER/Reflectance/Metadata/Spectral_Data/Wavelength")
Extract Z-dimension data slice
Next, we will extract all reflectance values for one pixel. This makes up the spectral signature or profile of the pixel. To do that, we'll use the h5read() function. Here we pick an arbitrary pixel at (100,35), and use the NULL value to select all bands from that location.
# extract all bands from a single pixel
aPixel <- h5read(h5_file,"/SJER/Reflectance/Reflectance_Data",index=list(NULL,100,35))
# The line above generates a vector of reflectance values.
# Next, we reshape the data and turn them into a dataframe
b <- adply(aPixel,c(1))
# create clean data frame
aPixeldf <- b[2]
# add wavelength data to matrix
aPixeldf$Wavelength <- wavelengths
head(aPixeldf)
## V1 Wavelength
## 1 206 381.6035
## 2 266 386.6132
## 3 274 391.6229
## 4 297 396.6327
## 5 236 401.6424
## 6 236 406.6522
Scale Factor
Then, we can pull the spatial attributes that we'll need to adjust the reflectance values. Often, large raster data contain floating point (values with decimals) information. However, floating point data consume more space (yield a larger file size) compared to integer values. Thus, to keep the file sizes smaller, the data will be scaled by a factor of 10, 100, 10000, etc. This scale factor will be noted in the data attributes.
# grab scale factor from the Reflectance attributes
reflectanceAttr <- h5readAttributes(h5_file,"/SJER/Reflectance/Reflectance_Data" )
scaleFact <- reflectanceAttr$Scale_Factor
# add scaled data column to DF
aPixeldf$scaled <- (aPixeldf$V1/as.vector(scaleFact))
# make nice column names
names(aPixeldf) <- c('Reflectance','Wavelength','ScaledReflectance')
head(aPixeldf)
## Reflectance Wavelength ScaledReflectance
## 1 206 381.6035 0.0206
## 2 266 386.6132 0.0266
## 3 274 391.6229 0.0274
## 4 297 396.6327 0.0297
## 5 236 401.6424 0.0236
## 6 236 406.6522 0.0236
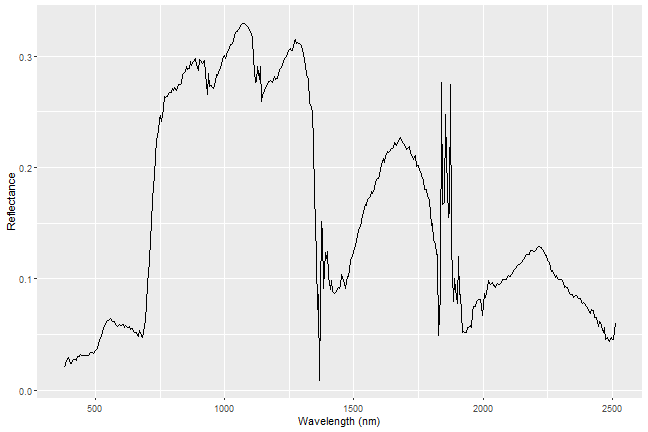
Plot Spectral Signature
Now we're ready to plot our spectral signature!
ggplot(data=aPixeldf)+
geom_line(aes(x=Wavelength, y=ScaledReflectance))+
xlab("Wavelength (nm)")+
ylab("Reflectance")
Get Lesson Code
Select pixels and compare spectral signatures in R
Authors: Donal O'Leary
Last Updated: Feb 19, 2024
In this tutorial, we will learn how to plot spectral signatures of several
different land cover types using an interactive click feature of the
terra package.
Learning Objectives
After completing this activity, you will be able to:
- Extract and plot spectra from an HDF5 file.
- Work with groups and datasets within an HDF5 file.
- Use the
terra::click()function to interact with an RGB raster image
Things You’ll Need To Complete This Tutorial
To complete this tutorial you will need the most current version of R and, preferably, RStudio loaded on your computer.
R Libraries to Install:
-
rhdf5:
install.packages("BiocManager"),BiocManager::install("rhdf5") -
terra:
install.packages('terra') -
plyr:
install.packages('plyr') -
reshape2:
install.packages('reshape2') -
ggplot2:
install.packages('ggplot2') -
neonUtilities:
install.packages('neonUtilities')
More on Packages in R - Adapted from Software Carpentry.
Data to Download
These hyperspectral remote sensing data provide information on the National Ecological Observatory Network's San Joaquin Experimental Range (SJER) field site in March of 2021. The data used in this lesson is the 1km by 1km mosaic tile named NEON_D17_SJER_DP3_257000_4112000_reflectance.h5. If you already completed the previous lesson in this tutorial series, you do not need to download this data again. The entire SJER reflectance dataset can be accessed from the NEON Data Portal.
Set Working Directory: This lesson assumes that you have set your working directory to the location of the downloaded data.
An overview of setting the working directory in R can be found here.
Recommended Skills
This tutorial will require that you be comfortable navigating HDF5 files, and have an understanding of what spectral signatures are. For additional information on these topics, we highly recommend you work through the earlier tutorials in this Introduction to Hyperspectral Remote Sensing Data series before starting on this tutorial.
Getting Started
First, we need to load our required packages and set the working directory.
# load required packages
library(rhdf5)
library(reshape2)
library(terra)
library(plyr)
library(ggplot2)
library(grDevices)
# set working directory, you can change this if desired
wd <- "~/data/"
setwd(wd)
Download the reflectance tile, if you haven't already, using neonUtilities::byTileAOP:
byTileAOP(dpID = 'DP3.30006.001',
site = 'SJER',
year = '2021',
easting = 257500,
northing = 4112500,
savepath = wd)
And then we can read in the hyperspectral hdf5 data. We will also collect a few other important pieces of information (band wavelengths and scaling factor) while we're at it.
# define filepath to the hyperspectral dataset
h5_file <- paste0(wd,"DP3.30006.001/neon-aop-products/2021/FullSite/D17/2021_SJER_5/L3/Spectrometer/Reflectance/NEON_D17_SJER_DP3_257000_4112000_reflectance.h5")
# read in the wavelength information from the HDF5 file
wavelengths <- h5read(h5_file,"/SJER/Reflectance/Metadata/Spectral_Data/Wavelength")
# grab scale factor from the Reflectance attributes
reflInfo <- h5readAttributes(h5_file,"/SJER/Reflectance/Reflectance_Data" )
scaleFact <- reflInfo$Scale_Factor
Now, we will read in the RGB image that we created in an earlier tutorial and plot it.
# read in RGB image as a 'stack' rather than a plain 'raster'
rgbStack <- rast(paste0(wd,"NEON_hyperspectral_tutorial_example_RGB_image.tif"))
# plot as RGB image, with a linear stretch
plotRGB(rgbStack,
r=1,g=2,b=3, scale=300,
stretch = "lin")
![]()
Interactive click Function from the terra Package
Next, we use an interactive clicking function to identify the pixels that we want to extract spectral signatures for. To follow along with this tutorial, we suggest the following six cover types (exact locations shown in the image below).
- Water
- Tree canopy (avoid the shaded northwestern side of the tree)
- Irrigated grass
- Bare soil (baseball diamond infield)
- Building roof (blue)
- Road
As shown here:

Data Tip: Note from the terra::click Description (which you can read by typing help("click"): click "does not work well on the default RStudio plotting device. To work around that, you can first run dev.new(noRStudioGD = TRUE) which will create a separate window for plotting, then use plot() followed by click() and click on the map."
For this next part, if you are following along in RStudio, you will need to enter these line below directly in the Console. dev.new(noRStudioGD = TRUE) will open up a separate window for plotting, which is where you will click the pixels to extract spectra, using the terra::click functionality.
dev.new(noRStudioGD = TRUE)
Now we can create our RGB plot, and start clicking on this in the pop-out Graphics window.
# change plotting parameters to better see the points and numbers generated from clicking
par(col="red", cex=2)
# use a histogram stretch in order to provide more contrast for selecting pixels
plotRGB(rgbStack, r=1, g=2, b=3, scale=300, stretch = "hist")
# use the 'click' function
c <- click(rgbStack, n = 6, id=TRUE, xy=TRUE, cell=TRUE, type="p", pch=16, col="red", col.lab="red")
Once you have clicked your six points, the graphics window should close. If you want to choose new points, or if you accidentally clicked a point that you didn't intend to, run the previous 2 chunks of code again to re-start.
The click() function identifies the cell number that you clicked, but in order to extract spectral signatures, we need to convert that cell number into a row and column, as shown here:
# convert raster cell number into row and column (used to extract spectral signature below)
c$row <- c$cell%/%nrow(rgbStack)+1 # add 1 because R is 1-indexed
c$col <- c$cell%%ncol(rgbStack)
Extract Spectral Signatures from HDF5 file
Next, we will loop through each of the cells that and use the h5read() function to extract the reflectance values of all bands from the given pixel (row and column).
# create a new dataframe from the band wavelengths so that we can add the reflectance values for each cover type
pixel_df <- as.data.frame(wavelengths)
# loop through each of the cells that we selected
for(i in 1:length(c$cell)){
# extract spectral values from a single pixel
aPixel <- h5read(h5_file,"/SJER/Reflectance/Reflectance_Data",
index=list(NULL,c$col[i],c$row[i]))
# scale reflectance values from 0-1
aPixel <- aPixel/as.vector(scaleFact)
# reshape the data and turn into dataframe
b <- adply(aPixel,c(1))
# rename the column that we just created
names(b)[2] <- paste0("Point_",i)
# add reflectance values for this pixel to our combined data.frame called pixel_df
pixel_df <- cbind(pixel_df,b[2])
}
Plot Spectral signatures using ggplot2
Finally, we have everything that we need to plot the spectral signatures for each of the pixels that we clicked. In order to color our lines by the different land cover types, we will first reshape our data using the melt() function, then plot the spectral signatures.
# Use the melt() function to reshape the dataframe into a format that ggplot prefers
pixel.melt <- reshape2::melt(pixel_df, id.vars = "wavelengths", value.name = "Reflectance")
# Now, let's plot the spectral signatures!
ggplot()+
geom_line(data = pixel.melt, mapping = aes(x=wavelengths, y=Reflectance, color=variable), lwd=1.5)+
scale_colour_manual(values = c("blue3","green4","green2","tan4","grey50","black"),
labels = c("Water","Tree","Grass","Soil","Roof","Road"))+
labs(color = "Cover Type")+
ggtitle("Land cover spectral signatures")+
theme(plot.title = element_text(hjust = 0.5, size=20))+
xlab("Wavelength")
![]()
Nice! However, there seems to be something weird going on in the wavelengths near ~1400nm and ~1850 nm...
Atmospheric Absorption Bands
Those irregularities around 1400nm and 1850 nm are two major atmospheric absorption bands - regions where gasses in the atmosphere (primarily carbon dioxide and water vapor) absorb radiation, and therefore, obscure the reflected radiation that the imaging spectrometer measures. Fortunately, the lower and upper bound of each of those atmospheric absorption bands is specified in the HDF5 file. Let's read those bands and plot rectangles where the reflectance measurements are obscured by atmospheric absorption.
# grab reflectance metadata (which contains absorption band limits)
reflMetadata <- h5readAttributes(h5_file,"/SJER/Reflectance" )
ab1 <- reflMetadata$Band_Window_1_Nanometers
ab2 <- reflMetadata$Band_Window_2_Nanometers
# Plot spectral signatures again with grey rectangles highlighting the absorption bands
ggplot()+
geom_line(data = pixel.melt, mapping = aes(x=wavelengths, y=Reflectance, color=variable), lwd=1.5)+
geom_rect(mapping=aes(ymin=min(pixel.melt$Reflectance),ymax=max(pixel.melt$Reflectance), xmin=ab1[1], xmax=ab1[2]), color="black", fill="grey40", alpha=0.8)+
geom_rect(mapping=aes(ymin=min(pixel.melt$Reflectance),ymax=max(pixel.melt$Reflectance), xmin=ab2[1], xmax=ab2[2]), color="black", fill="grey40", alpha=0.8)+
scale_colour_manual(values = c("blue3","green4","green2","tan4","grey50","black"),
labels = c("Water","Tree","Grass","Soil","Roof","Road"))+
labs(color = "Cover Type")+
ggtitle("Land cover spectral signatures with\n atmospheric absorption bands greyed out")+
theme(plot.title = element_text(hjust = 0.5, size=20))+
xlab("Wavelength")
![]()
Now we can clearly see that the noisy sections of each spectral signature are within the atmospheric absorption bands. For our final step, let's take all reflectance values from within each absorption band and set them to NA to remove the noisiest sections from the plot.
# Duplicate the spectral signatures into a new data.frame
pixel.melt.masked <- pixel.melt
# Mask out all values within each of the two atmospheric absorption bands
pixel.melt.masked[pixel.melt.masked$wavelengths>ab1[1]&pixel.melt.masked$wavelengths<ab1[2],]$Reflectance <- NA
pixel.melt.masked[pixel.melt.masked$wavelengths>ab2[1]&pixel.melt.masked$wavelengths<ab2[2],]$Reflectance <- NA
# Plot the masked spectral signatures
ggplot()+
geom_line(data = pixel.melt.masked, mapping = aes(x=wavelengths, y=Reflectance, color=variable), lwd=1.5)+
scale_colour_manual(values = c("blue3","green4","green2","tan4","grey50","black"),
labels = c("Water","Tree","Grass","Soil","Roof","Road"))+
labs(color = "Cover Type")+
ggtitle("Land cover spectral signatures with\n atmospheric absorption bands removed")+
theme(plot.title = element_text(hjust = 0.5, size=20))+
xlab("Wavelength")
![]()
There you have it, spectral signatures for six different land cover types, with the regions from the atmospheric absorption bands removed.
Challenge: Compare Spectral Signatures
There are many interesting comparisons to make with spectral signatures. Try these challenges to explore hyperspectral data further:
-
Compare six different types of vegetation, and pick an appropriate color for each of their lines. A nice guide to the many different color options in R can be found here.
-
What happens if you only click five points? What about ten? How does this change the spectral signature plots, and can you fix any errors that occur?
-
Does shallow water have a different spectral signature than deep water?