Workshop
NEON Data Institute 2016: Remote Sensing with Reproducible Workflows in R
NEON
-
NEON's Data Institutes provide critical skills and foundational knowledge for graduate students and early career scientists working with heterogeneous spatio-temporal data to address ecological questions.
Data Institute Overview
Our 2016 Institute focused on remote sensing of vegetation using open source tools and reproducible science workflows. The programming language of instruction in 2016 was R. This Institute was held at NEON headquarters in June 2016.
In addition to the six day institute there were three weeks of pre-institute materials is to ensure that everyone comes to the Institute ready to work in a collaborative research environment. Pre-institute materials are online & individually paced, expect to spend 1-5 hrs/week depending on familiarity with the topic.
Schedule
| Time | Day | Description |
|---|---|---|
| -- | Computer Setup Materials | |
| -- | 25 May - 1 June | Intro to NEON & Reproducible Science |
| -- | 2-8 June | Version Control & Collaborative Science with Git & GitHub |
| -- | 9-15 June | Documentation of Your Workflow with R Markdown |
| -- | 19-24 June | Data Institute |
| 7:50am - 10:00 pm | Monday | Intro to NEON, Intro to HDF5 & Hyperspectral Remote Sensing |
| 8:00am - 6:30pm | Tuesday | Intro to LiDAR data, Automated Workflows |
| 8:00am - 6:30pm | Wednesday | Remote Sensing Uncertainty |
| 8:00am - 6:30pm | Thursday | LiDAR & Hyperspectral Data Fusion |
| 9:00am - 6:30pm | Friday | Individual/Group Applications |
| 9:00am - 2:00pm | Saturday | Group Application Presentations |
Key Dates
- Application Deadline: March 28, 2016
- Notification of Acceptance: April 4, 2016
- Tuition payment due by: April 18, 2016
- Pre-institute online activities: June 1-17, 2016
- Institute Dates: June 20-25, 2016
Instructors
Dr. Leah Wasser, Supervising Scientist, NEON: As part of her work at NEON, Leah is passionate about helping the scientific community harness the power of remote sensing and other large spatio-temporal data using efficient, quantitative, reproducible approaches and open science workflows to better understand ecological change over time. Leah has a Ph.D. in ecology with a focus on using remote sensing techniques to measure landscape level ecological change.
Dr. Naupaka Zimmerman, Assistant Professor of Biology, University of San Francisco: Naupaka’s research focuses on the microbial ecology of plant-fungal interactions. Naupaka brings to the course experience and enthusiasm for reproducible workflows developed after discovering how challenging it is to keep track of complex analyses in his own dissertation and postdoctoral work. As a co-founder of the International Network of Next-Generation Ecologists and an instructor and lesson maintainer for Software Carpentry and Data Carpentry, Naupaka is very interested in providing and improving training experiences in open science and reproducible research methods.
Dr. Kyla Dahlin, Assistant Professor, Michigan State University: Kyla's research aims to better understand and quantify ecosystem processes and disturbance responses through the application of emerging technologies, including air- and space-borne remote sensing, spatial statistics, and process-based modeling. She is currently interested in semi-arid forest/grassland transition zones, where vegetation patterns are readily observable but poorly understood. Kyla approaches questions by integrating observational data, modeling, and focused field experiments to both refine our understanding of ecosystem function and to improve our ability to predict how ecosystems and the climate will change in the future.
Online Resources
The teaching materials from the 2016 Data Institute are provided free on this site for use outside the Data Institute. They can be found in the Workshop Materials section of this page. These materials were designed to be used in the context of the workshop with an instructor, however, they may also be suitable for self-paced online instruction.
You too can watch several of the presentations that were given at the 2016 Data Institute!
- Big Data, Open Data and Biodiversity with David Schimel
- An Introduction to Hyperspectral Remote Sensing
- An Introduction to Full Waveform LiDAR
- NEON Remote Sensing Vegetation Indices, Data Products & Uncertainty Measurements
- NEON Terrestrial Observation Vegetation Sampling
2016 Data Institute Recap
In addition to the three core faculty listed above the Data Institute participants were instructed by and interacted with guest instructors and NEON project scientists:
- Lindsay Powers, H5 Group – HDF5 data structure
- Chris Crosby, UNAVCO/Open Topography – LiDAR remote sensing
- David Schimel, NASA Jet Propulsion Lab – remote sensing, open science, ecology
- David Hulslander, NEON – Remote sensing data processing
- Tristan Goulden, NEON – Remote sensing theory & Hyperspectral remote sensing
- Nathan Leisso, NEON – Introduction to NEON AOP data collection and processing
- Courtney Meier, NEON – NEON in situ field measurements.
- Keith Krause, NEON – NEON full waveform LiDAR
Participants
Participants came from institutions in the USA, Canada and the Netherlands. While 70% of the participants were graduate students, the Data Institute also attracted an undergraduate student, post-docs, and university research staff and faculty.
[[nid:6008 align=left&size=medium]]
Participant were interested in using remote sensing data to answer a wide range of questions from wanting to be able to characterize forest structure and composition to using time series to detect vegetation disturbance patterns to from remote sensing data.
According to NEON science educator, Megan Jones, “Participants really appreciated the opportunities to work with data in small-group settings and the emphasis of using reproducible science methods. The science theme for 2016 was use of remote sensing data, but this was taught along with reproducible science methods including the importance of well documented code, version control and collaborative tools like GitHub, and quick sharing of results using RMarkdown and knitr.”
Institute outcomes
At the end of the Institute, participants presented group projects illustrating the use of reproducible workflows with remote sensing data. The skills learned are applicable to remote sensing data from any source, however, all participants were allowed to use NEON remote sensing data as well as their own data sets. According to Robert Paul from the University of Illinois at Urbana-Champaign, “The course offered a comprehensive overview of best practices for managing and analyzing remote sensing data, and how to make data analysis workflows well-documented, collaborative, and reproducible.”
Sarah Graves, from the University of Florida, said, “The NEON Data Institute gave us the tools to work with novel ecological data. With our own knowledge of the domain combined with NEON data and tools, we are in a position to ask novel ecological questions that will advance the field of ecology beyond what has been traditionally possible.” Jeff Atkins of Virginia Commonwealth University added, “Ecology increasingly depends on "big data" and remote sensing and scientists need the skills necessary to work with this data and to inform their hypotheses. NEON does an amazing job at helping scientists learn how to work with and use a suite of data and data products.”
Group projects
Exploring the relationship between functional traits and spectral reflectance for Ordway Swisher Biological Station, FL
Sarah Graves, Jeff Atkins, Kunxuan Wang, and Catherine Hulshof de la Pena
We calculated plot-level foliar nitrogen content and functional diversity from in situ data. These metrics were related to mean plot reflectance and a spectral diversity metric from a PCA transformation.
Describing landscape-level phenology with MODIS vegetation index time series
Robert Paul, Jeff Stephens
This workflow detects the length of time for NDVI and EVI to go from baseline to peak over the course of the year. Each pixel is classified with a value reflecting the length of time in the year for NDVI and EVI to reach peak greenness.
Characterizing the forest using trees: how do forest characteristics vary with respect to disturbance history at Soaproot Saddle
We attempted species-level classification using Random Forest on LiDAR and imaging spectroscopy.
Megan Cattau, Stella Cousins, Kristin Braziunas, Allie Weill
Towards individual tree crown segmentation with spectral indices
Enrique Montano & Dave McCaffrey
We attempted to implement an individual tree crown extraction algorithm, optimized with vegetation structure data from in situ plots. The ability to identify individual tree canopy with confidence will allow for comparison of spectral indices among individuals and across species.
Plant structure and function in complex terrain: Landscape controls and microclimatic consequences
Holly Andrews, Nate Looker, Amy Hudson
We examined climate, topography, and vegetation interactions. Specifically, we assessed spectral and LiDAR-based properties of vegetation across topographic gradients of water availability and compared land surface temperature to NDVI.
Upscaling Structure for Soaproot Field Site, California
Cassondra Walker, Jon Weiner, Richard Remigio
We attempted to link vegetation indices to plot-level tree characteristics, and then upscale those indices to the landscape scale to predict structure that was derived from LiDAR.
Using HyperSpectral Imaging techniques to predict foliar nutrient concentrations
Michiel Veldhuis
This page includes all of the materials needed for the Data Institute including the pre-institute materials. Please use the sidebar menu to find the appropriate week or day. If you have problems with any of the materials please email us or use the comments section at the bottom of the appropriate page.
Please note that the format of the HDF5 files use for many of the tutorials for the 2016 Data Institute are in a format no longer used for NEON data. We are in the process of updating the content to the new HDF5 format. As the tutorials are updated they will be linked below. In the meantime, you can view the tutorials using the old format on the original NEON Data Institute 2016 website.
Pre-Institute: Computer Set Up Materials
It is important that you have your computer setup, prior to diving into the pre-institute materials in week 2!
Please review the links below to setup the laptop you will be bringing to the Data Institute.
Let's Get Your Computer Setup!
Go to each of the following tutorials and complete the directions to set your computer up for the Data Institute.
Install Git, Bash Shell, R & RStudio
This page outlines the tools and resources that you will need to get started working on the many R-based tutorials that NEON provides.
Checklist
This checklist includes the tools that need to be set-up on your computer. Detailed directions to accomplish each objective are below.
- Install Bash shell (or shell of preference)
- Install Git
- Install R & RStudio
Bash/Shell Setup
Install Bash for Windows
- Download the Git for Windows installer.
- Run the installer and follow the steps below (these may look slightly different depending on Git version number):
- Welcome to the Git Setup Wizard: Click on "Next".
- Information: Click on "Next".
- Select Destination Location: Click on "Next".
- Select Components: Click on "Next".
- Select Start Menu Folder: Click on "Next".
- Adjusting your PATH environment: Select "Use Git from the Windows Command Prompt" and click on "Next". If you forgot to do this programs that you need for the event will not work properly. If this happens rerun the installer and select the appropriate option.
- Configuring the line ending conversions: Click on "Next". Keep "Checkout Windows-style, commit Unix-style line endings" selected.
- Configuring the terminal emulator to use with Git Bash: Select "Use Windows' default console window" and click on "Next".
- Configuring experimental performance tweaks: Click on "Next".
- Completing the Git Setup Wizard: Click on "Finish".
This will provide you with both Git and Bash in the Git Bash program.
Install Bash for Mac OS X
The default shell in all versions of Mac OS X is bash, so no
need to install anything. You access bash from the Terminal
(found in
/Applications/Utilities). You may want to keep
Terminal in your dock for this workshop.
Install Bash for Linux
The default shell is usually Bash, but if your
machine is set up differently you can run it by opening a
terminal and typing bash. There is no need to
install anything.
Git Setup
Git is a version control system that lets you track who made changes to what when and has options for easily updating a shared or public version of your code on GitHub. You will need a supported web browser (current versions of Chrome, Firefox or Safari, or Internet Explorer version 9 or above).
Git installation instructions borrowed and modified from Software Carpentry.
Git for Windows
Git should be installed on your computer as part of your Bash install.Git on Mac OS X
Video Tutorial
Install Git on Macs by downloading and running the most recent installer for
"mavericks" if you are using OS X 10.9 and higher -or- if using an
earlier OS X, choose the most recent "snow leopard" installer, from
this list.
After installing Git, there will not be anything in your
/Applications folder, as Git is a command line program.
Git on Linux
If Git is not already available on your machine you can try to
install it via your distro's package manager. For Debian/Ubuntu run
sudo apt-get install git and for Fedora run
sudo yum install git.
Setting Up R & RStudio
Windows R/RStudio Setup
- Please visit the CRAN Website to download the latest version of R for windows.
- Run the .exe file that was just downloaded
- Go to the RStudio Download page
- Download the latest version of Rstudio for Windows
- Double click the file to install it
Once R and RStudio are installed, click to open RStudio. If you don't get any error messages you are set. If there is an error message, you will need to re-install the program.
Mac R/RStudio Setup
- Go to CRAN and click on Download R for (Mac) OS X
- Select the .pkg file for the version of OS X that you have and the file will download.
- Double click on the file that was downloaded and R will install
- Go to the RStudio Download page
- Download the latest version of Rstudio for Mac
- Once it's downloaded, double click the file to install it
Once R and RStudio are installed, click to open RStudio. If you don't get any error messages you are set. If there is an error message, you will need to re-install the program.
Linux R/RStudio Setup
- R is available through most Linux package managers.
You can download the binary files for your distribution
from CRAN. Or
you can use your package manager (e.g. for Debian/Ubuntu
run
sudo apt-get install r-baseand for Fedora runsudo yum install R). - To install RStudio, go to the RStudio Download page
- Under Installers select the version for your distribution.
- Once it's downloaded, double click the file to install it
Once R and RStudio are installed, click to open RStudio. If you don't get any error messages you are set. If there is an error message, you will need to re-install the program.
Set up GitHub Working Directory - Quick Intro to Bash
Checklist
Once you have Git and Bash installed, you are ready to configure Git.
On this page you will:
- Create a directory for all future GitHub repositories created on your computer
To ensure Git is properly installed and to create a working directory for GitHub, you will need to know a bit of shell -- brief crash course below.
Crash Course on Shell
The Unix shell has been around longer than most of its users have been alive. It has survived so long because it’s a power tool that allows people to do complex things with just a few keystrokes. More importantly, it helps them combine existing programs in new ways and automate repetitive tasks so they aren’t typing the same things over and over again. Use of the shell is fundamental to using a wide range of other powerful tools and computing resources (including “high-performance computing” supercomputers).
This section is an abbreviated form of Software Carpentry’s The Unix Shell for Novice’s workshop lesson series. Content and wording (including all the above) is heavily copied and credit is due to those creators (full author list).
Our goal with shell is to:
- Set up the directory where we will store all of the GitHub repositories during the Institute,
- Make sure Git is installed correctly, and
- Gain comfort using bash so that we can use it to work with Git & GitHub.
Accessing Shell
How one accesses the shell depends on the operating system being used.
- OS X: The bash program is called Terminal. You can search for it in Spotlight.
- Windows: Git Bash came with your download of Git for Windows. Search Git Bash.
- Linux: Default is usually bash, if not, type
bashin the terminal.
Bash Commands
$
The dollar sign is a prompt, which shows us that the shell is waiting for input; your shell may use a different character as a prompt and may add information before the prompt.
When typing commands, either from these tutorials or from other sources, do not
type the prompt ($), only the commands that follow it.
In these tutorials, subsequent lines that follow a prompt and do not start with
$ are the output of the command.
listing contents - ls
Next, let's find out where we are by running a command called pwd -- print
working directory. At any moment, our current working directory is our
current default directory. I.e., the directory that the computer assumes we
want to run commands in unless we explicitly specify something else. Here, the
computer's response is /Users/neon, which is NEON’s home directory:
$ pwd
/Users/neon
If you are not, by default, in your home directory, you get there by typing:
$ cd ~
Now let's learn the command that will let us see the contents of our own
file system. We can see what's in our home directory by running ls --listing.
$ ls
Applications Documents Library Music Public
Desktop Downloads Movies Pictures
(Again, your results may be slightly different depending on your operating system and how you have customized your filesystem.)
ls prints the names of the files and directories in the current directory in
alphabetical order, arranged neatly into columns.
Change directory -- cd
Now we want to move into our Documents directory where we will create a
directory to host our GitHub repository (to be created in Week 2). The command
to change locations is cd followed by a directory name if it is a
sub-directory in our current working directory or a file path if not.
cd stands for "change directory", which is a bit misleading: the command
doesn't change the directory, it changes the shell's idea of what directory we
are in.
To move to the Documents directory, we can use the following series of commands to get there:
$ cd Documents
These commands will move us from our home directory into our Documents
directory. cd doesn't print anything, but if we run pwd after it, we can
see that we are now in /Users/neon/Documents.
If we run ls now, it lists the contents of /Users/neon/Documents, because
that's where we now are:
$ pwd
/Users/neon/Documents
$ ls
data/ elements/ animals.txt planets.txt sunspot.txt
To use cd, you need to be familiar with paths, if not, read the section on
Full, Base, and Relative Paths .
Make a directory -- mkdir
Now we can create a new directory called GitHub that will contain our GitHub
repositories when we create them later.
We can use the command mkdir NAME-- “make directory”
$ mkdir GitHub
There is not output.
Since GitHub is a relative path (i.e., doesn't have a leading slash), the
new directory is created in the current working directory:
$ ls
data/ elements/ GitHub/ animals.txt planets.txt sunspot.txt
Is Git Installed Correctly?
All of the above commands are bash commands, not Git specific commands. We still need to check to make sure git installed correctly. One of the easiest ways is to check to see which version of git we have installed.
Git commands start with git.
We can use git --version to see which version of Git is installed
$ git --version
git version 2.5.4 (Apple Git-61)
If you get a git version number, then Git is installed!
If you get an error, Git isn’t installed correctly. Reinstall and repeat.
Setup Git Global Configurations
Now that we know Git is correctly installed, we can get it set up to work with.
The text below is modified slightly from Software Carpentry's Setting up Git lesson.
When we use Git on a new computer for the first time, we need to configure a few things. Below are a few examples of configurations we will set as we get started with Git:
- our name and email address,
- to colorize our output,
- what our preferred text editor is,
- and that we want to use these settings globally (i.e. for every project)
On a command line, Git commands are written as git verb, where verb is what
we actually want to do.
Set up you own git with the following command, using your own information instead of NEON's.
$ git config --global user.name "NEON Science"
$ git config --global user.email "neon@BattelleEcology.org"
$ git config --global color.ui "auto"
Then set up your favorite text editor following this table:
| Editor | Configuration command |
|---|---|
| nano | $ git config --global core.editor "nano -w" |
| Text Wrangler | $ git config --global core.editor "edit -w" |
| Sublime Text (Mac) | $ git config --global core.editor "subl -n -w" |
| Sublime Text (Win, 32-bit install) | $ git config --global core.editor "'c:/program files (x86)/sublime text 3/sublime_text.exe' -w" |
| Sublime Text (Win, 64-bit install) | $ git config --global core.editor "'c:/program files/sublime text 3/sublime_text.exe' -w" |
| Notepad++ (Win) | $ git config --global core.editor "'c:/program files (x86)/Notepad++/notepad++.exe' -multiInst -notabbar -nosession -noPlugin" |
| Kate (Linux) | $ git config --global core.editor "kate" |
| Gedit (Linux) | $ git config --global core.editor "gedit -s -w" |
| emacs | $ git config --global core.editor "emacs" |
| vim | $ git config --global core.editor "vim" |
The four commands we just ran above only need to be run once:
the flag --global tells Git to use the settings for every project in your user
account on this computer.
You can check your settings at any time:
$ git config --list
You can change your configuration as many times as you want; just use the same commands to choose another editor or update your email address.
Now that Git is set up, you will be ready to start the Week 2 materials to learn about version control and how Git & GitHub work.
Data Institute: Install Required R Packages
R and RStudio
Once R and RStudio are installed (in Install Git, Bash Shell, R & RStudio ), open RStudio to make sure it works and you don’t get any error messages. Then, install the needed R packages.
Install/Update R Packages
Please make sure all of these packages are installed and up to date on your computer prior to the Institute.
-
install.packages(c("raster", "rasterVis", "rgdal", "rgeos", "rmarkdown","knitr", "plyr", "dplyr", "ggplot2", "plotly")) - The
rhdf5package is not on CRAN and must be downloaded directly from Bioconductor. The can be done using these two commands directly in your R console.-
#install.packages("BiocManager") -
#BiocManager::install("rhdf5")
-
Install QGIS & HDF5View
Install HDFView
The free HDFView application allows you to explore the contents of an HDF5 file.
To install HDFView:
-
Click to go to the download page.
-
From the section titled HDF-Java 2.1x Pre-Built Binary Distributions select the HDFView download option that matches the operating system and computer setup (32 bit vs 64 bit) that you have. The download will start automatically.
-
Open the downloaded file.
- Mac - You may want to add the HDFView application to your Applications directory.
- Windows - Unzip the file, open the folder, run the .exe file, and follow directions to complete installation.
- Open HDFView to ensure that the program installed correctly.
Install QGIS
QGIS is a free, open-source GIS program. Installation is optional for the 2018 Data Institute. We will not directly be working with QGIS, however, some past participants have found it useful to have during the capstone projects.
To install QGIS:
Download the QGIS installer on the QGIS download page here. Follow the installation directions below for your operating system.
Windows
- Select the appropriate QGIS Standalone Installer Version for your computer.
- The download will automatically start.
- Open the .exe file and follow prompts to install (installation may take a while).
- Open QGIS to ensure that it is properly downloaded and installed.
Mac OS X
- Select KyngChaos QGIS download page. This will take you to a new page.
- Select the current version of QGIS. The file download (.dmg format) should start automatically.
- Once downloaded, run the .dmg file. When you run the .dmg, it will create a directory of installer packages that you need to run in a particular order. IMPORTANT: read the READ ME BEFORE INSTALLING.rtf file!
Install the packages in the directory in the order indicated.
- GDAL Complete.pkg
- NumPy.pkg
- matplotlib.pkg
- QGIS.pkg - NOTE: you need to install GDAL, NumPy and matplotlib in order to successfully install QGIS on your Mac!
Once all of the packages are installed, open QGIS to ensure that it is properly installed.
LINUX
- Select the appropriate download for your computer system.
- Note: if you have previous versions of QGIS installed on your system, you may run into problems. Check out
Pre-Institute Week 1: Introduction to NEON & Reproducible Science
In the first week of the pre-institute activities, we will review the NEON project. We will also provide you with a general overview of reproducible science. Over the next few weeks will we ask you to review materials and submit something that demonstrates you have mastered the materials.
Learning Objectives
After completing these activities, you will be able to:
- Explain sources of uncertainty in remote sensing data.
- Measure the differences between a metric derived from remote sensing data and the same metric derived from data collected on the ground.
Week 1 Assignment
After reviewing the materials below, please write up a summary of a project that you are interested working on at the Data Institute. Be sure to consider what data you will need (NEON or other). You will have time to refine your idea over the next few weeks. Save this document as you will submit it next week as a part of week 2 materials!
Deadline: Please complete this by Thursday June 2nd 2016 @ 11:59 MDT.
Week 1 Materials
Please carefully read and review the materials below:
Introduction to the National Ecological Observatory Network (NEON)
Here we will provide an overview of the National Ecological Observatory Network (NEON). Please carefully read through these materials and links that discuss NEON’s mission and design.
Learning Objectives
At the end of this activity, you will be able to:
- Explain the mission of the National Ecological Observatory Network (NEON).
- Explain the how sites are located within the NEON project design.
- Explain the different types of data that will be collected and provided by NEON.
The NEON Project Mission & Design
To capture ecological heterogeneity across the United States, NEON’s design divides the continent into 20 statistically different eco-climatic domains. Each NEON field site is located within an eco-climatic domain.
The Science and Design of NEON
To gain a better understanding of the broad scope fo NEON watch this 4 minute long video.
Please, read the following page about NEON's mission.
Data Institute Participants -- Thought Question: How might/does the NEON project intersect with your current research or future career goals?
NEON's Spatial Design
The Spatial Design of NEON
Watch this 4:22 minute video exploring the spatial design of NEON field sites.
Please read the following page about NEON's Spatial Design:
Read this primer on NEON's Sampling Design
Read about the different types of field sites - core and relocatable
NEON Field Site Locations
Explore the NEON Field Site map taking note of the locations of
- Aquatic & terrestrial field sites.
- Core & relocatable field sites.
Explore the NEON field site map. Do the following:
- Zoom in on a study area of interest to see if there are any NEON field sites that are nearby.
- Use the menu below the map to filter sites by name, type, domain, or state.
- Select one field site of interest.
- Click on the marker in the map.
- Then click on Site Details to jump to the field site landing page.
Data Institute Participant -- Thought Questions: Use the map above to answer these questions. Consider the research question that you may explore as your Capstone Project at the Institute or about a current project that you are working on and answer the following questions:
- Are there NEON field sites that are in study regions of interest to you?
- What domains are the sites located in?
- What NEON field sites do your current research or Capstone Project ideas coincide with?
- Is the site(s) core or relocatable?
- Is it/are they terrestrial or aquatic?
- Are there data available for the NEON field site(s) that you are most interested in? What kind of data are available?
Data Tip: You can download maps, kmz, or shapefiles of the field sites here.
NEON Data
How NEON Collects Data
Watch this 3:06 minute video exploring the data that NEON collects.
Read the Data Collection Methods page to learn more about the different types of data that NEON collects and provides. Then, follow the links below to learn more about each collection method:
- Aquatic Observation System (AOS)
- Aquatic Instrument System (AIS)
- Terrestrial Instrument System (TIS) -- Flux Tower
- Terrestrial Instrument System (TIS) -- Soil Sensors and Measurements
- Terrestrial Organismal System (TOS)
- Airborne Observation Platform (AOP)
All data collection protocols and processing documents are publicly available. Read more about the standardized protocols and how to access these documents.
Specimens & Samples
NEON also collects samples and specimens from which the other data products are based. These samples are also available for research and education purposes. Learn more: NEON Biorepository.
Airborne Remote Sensing
Watch this 5 minute video to better understand the NEON Airborne Observation Platform (AOP).
Data Institute Participant – Thought Questions: Consider either your current or future research or the question you’d like to address at the Institute.
- Which types of NEON data may be more useful to address these questions?
- What non-NEON data resources could be combined with NEON data to help address your question?
- What challenges, if any, could you foresee when beginning to work with these data?
Data Tip: NEON also provides support to your own research including proposals to fly the AOP over other study sites, a mobile tower/instrumentation setup and others. Learn more here the Assignable Assets programs .
Access NEON Data
NEON data are processed and go through quality assurance quality control checks at NEON headquarters in Boulder, CO. NEON carefully documents every aspect of sampling design, data collection, processing and delivery. This documentation is freely available through the NEON data portal.
- Visit the NEON Data Portal - data.neonscience.org
- Read more about the quality assurance and quality control processes for NEON data and how the data are processed from raw data to higher level data products.
- Explore NEON Data Products. On the page for each data product in the catalog you can find the basic information about the product, find the data collection and processing protocols, and link directly to downloading the data.
- Additionally, some types of NEON data are also available through the data portals of other organizations. For example, NEON Terrestrial Insect DNA Barcoding Data is available through the Barcode of Life Datasystem (BOLD). Or NEON phenocam images are available from the Phenocam network site. More details on where else the data are available from can be found in the Availability and Download section on the Product Details page for each data product (visit Explore Data Products to access individual Product Details pages).
Pathways to access NEON Data
There are several ways to access data from NEON:
- Via the NEON data portal. Explore and download data. Note that much of the tabular data is available in zipped .csv files for each month and site of interest. To combine these files, use the neonUtilities package (R tutorial, Python tutorial).
- Use R or Python to programmatically access the data. NEON and community members have created code packages to directly access the data through an API. Learn more about the available resources by reading the Code Resources page or visiting the NEONScience GitHub repo.
- Using the NEON API. Access NEON data directly using a custom API call.
- Access NEON data through partner's portals. Where NEON data directly overlap with other community resources, NEON data can be accessed through the portals. Examples include Phenocam, BOLD, Ameriflux, and others. You can learn more in the documentation for individual data products.
Data Institute Participant – Thought Questions: Use the Data Portal tools to investigate the data availability for the field sites you’ve already identified in the previous Thought Questions.
- What types of aquatic/terrestrial data are currently available? Remote sensing data?
- Of these, what type of data are you most interested in working with for your project while at the Institute.
- For what time period does the data cover?
- What format is the downloadable file available in?
- Where is the metadata to support this data?
Data Institute Participants: Intro to NEON Culmination Activity
Write up a brief summary of a project that you might want to explore while at the Data Institute in Boulder, CO. Include the types of NEON (and other data) that you will need to implement this project. Save this summary as you will be refining and adding to your ideas over the next few weeks.
The goal of this activity if for you to begin to think about a Capstone Project that you wish to work on at the end of the Data Institute. This project will ideally be performed in groups, so over the next few weeks you'll have a chance to view the other project proposals and merge projects to collaborate with your colleagues.
The Importance of Reproducible Science
Verifiability and reproducibility are among the cornerstones of the scientific process. They are what allows scientists to "stand on the shoulder of giants". Maintaining reproducibility requires that all data management, analysis, and visualization steps behind the results presented in a paper are documented and available in full detail. Reproducibility here means that someone else should either be able to obtain the same results given all the documented inputs and the published instructions for processing them, or if not, the reasons why should be apparent. From Reproducible Science Curriculum
- Summarize the four facets of reproducibility.
- Describe several ways that reproducible workflows can improve your workflow and research.
- Explain several ways you can incorporate reproducible science techniques into your own research.
Getting Started with Reproducible Science
Please view the online slide-show below which summarizes concepts taught in the Reproducible Science Curriculum.
View Reproducible Science Slideshow
A Gap In Understanding

Reproducibility and Your Research
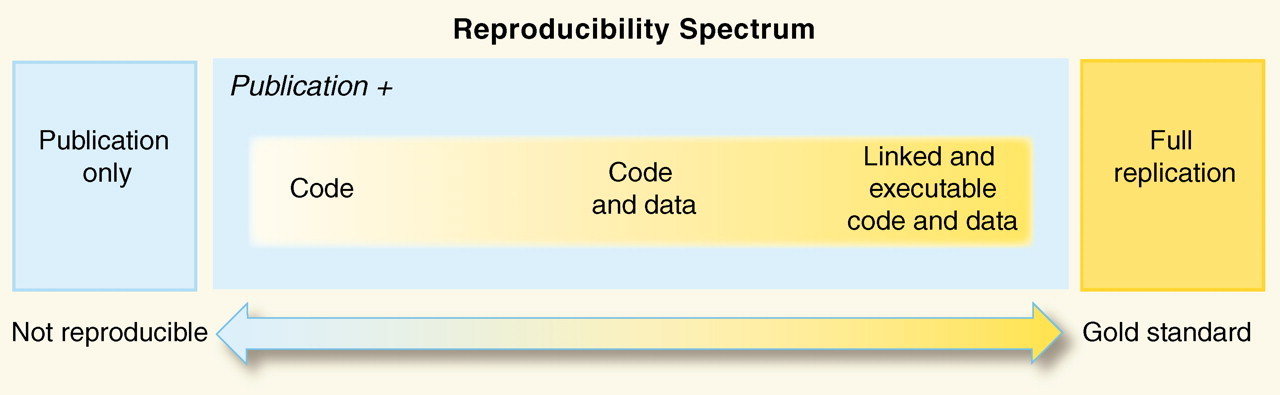
How reproducible is your current research?
View Reproducible Science Checklist
- Do you currently apply any of the items in the checklist to your research?
- Are there elements in the list that you are interested in incorporating into your workflow? If so, which ones?
Additional Readings (optional)
- Nature has collated and published (with open-access) a special archive on the Challenges of Irreproducible Science .
- The Nature Publishing group has also created a Reporting Checklist for its authors that focuses primaily on reporting issues but also includes sections for sharing code.
- Recent open-access issue of Ecography focusing on reproducible ecology and software packages available for use.
- A nice short blog post with an annotated bibliography of "Top 10 papers discussing reproducible research in computational science" from Lorena Barba: Barba group reproducibility syllabus.
Pre-Institute Week 2: Version Control & Collaborative Science with Git & Git Hub
The goal of the pre-institute materials is to ensure that everyone comes to the Institute ready to work in a collaborative research environment. If you recall, from last week, the four facets of reproducibility are documentation, organization, automation, and dissemination.
This week we will focus on learning to use tools to help us with these facets: Git and GitHub. The Git Hub environment supports both a collaborative approach to science through code sharing and dissemination, and a powerful version control system that supports both efficient project organization, and an effective way to save your work.
Learning Objectives
After completing these activities, you will be able to:
- Summarize the key components of a version control system
- Know how to setup a GitHub account
- Know how to setup Git locally
- Work in a collaborative workflow on GitHub
Week 2 Assignment
The assignment for this week is to revise the Data Institute capstone project summary that you developed last week. You will submit your project summary, with a brief biography to introduce yourself, to a shared GitHub repository.
Please complete this assignment by Thursday June 9th @ 11:59 PM MDT.
If you are familiar with forked repos and pull requests GitHub, and the use of Git in the command line, you may be able to complete the assignment without viewing the
tutorials.
Assignment: Version Control with GitHub
DUE: 21 June 2018
During the NEON Data Institute, you will share the code that you create daily
with everyone on the NEONScience/DI-NEON-participants repo.
Through this week’s tutorials, you have learned the basic skills needed to successfully share your work at the Institute including how to:
- Create your own GitHub user account,
- Set up Git on your computer (please do this on the computer you will be bringing to the Institute), and
- Create a Markdown file with a biography of yourself and the project you are interested in working on at the Institute. This biography was shared with the group via the Data Institute’s GitHub repo.
Checklist for this week’s Assignment:
You should have completed the following after Pre-institute week 2:
- Fork & clone the NEON-DataSkills/DI-NEON-participants repo.
- Create a .md file in the
participants/2018-RemoteSensing/pre-institute2-gitdirectory of the repo. Name the document LastName-FirstName.md. - Write a biography that introduces yourself to the other participants. Please
provide basic information including:
- name,
- domain of interest,
- one goal for the course,
- an updated version of your Capstone Project idea,
- and the list of data (NEON or other) to support the project that you created during last week’s materials.
- Push the document from your local computer to your GithHub repo.
- Created a Pull Request to merge this document back into the NEON-DataSkills/DI-NEON-participants repo.
NOTE: The Data Institute repository is a public repository, so all members of the Institute, as well as anyone in the general public who stumbles on the repo, can see the information. If you prefer not to share this information publicly, please submit the same document but use a pseudonym (cartoon character names would work well) and email us with the pseudonym so that we can connect the submitted document to you.
Have questions? No problem. Leave your question in the comment box below. It's likely some of your colleagues have the same question, too! And also likely someone else knows the answer.
Week 2 Materials
Please complete each of the short tutorials in this series.
Version Control with GitHub
Pre-Institute Week 3: Documentation of Your Workflow with R Markdown
In week 3, you will use the R Markdown file format to document code and efficiently publish code results & outputs. You will practice your Git skills by publishing your work in the NEON-WorkWithData/DI-NEON-participants GitHub repository.
Learning Objectives
After completing these activities, you will be able to:
- Use R Markdown and knitr to create code with formatted context text
- Describe the value of documented workflows
Week 3 Assignment
Please complete the activity and submit your work to the GitHub repo by 11:59 Thursday June 16th.
If you are familiar with using R Markdown files to document your workflow and knitting to HTML then you may be able to complete the assignment without viewing the tutorials.
Week 3 Materials
Please complete each of the short tutorials in this series:
Document Your Code with R Markdown
Monday: NEON, HDF5, & Hyperspectral Data
Welcome to Day One of the Institute!
After completing these activities, you will be able to:
- Describe the NEON project & NEON AOP data
- Understand key remote sensing data types (active vs passive sensors)
- Open and work with raster data stored in HDF5 format in R
- Explain the key components of the HDF5 data structure (groups, datasets and attributes)
- Open and use attribute data (metadata) from an HDF5 file in R
| Time | Topic | Instructor |
|---|---|---|
| 8:00 | Welcome & Introductions | |
| 9:30 | Introduction to NEON AOP | Nathan Leisso |
| 10:15 | Break | |
| 10:30 | Big Data, Open Data and Biodiversity (video) | Dave Schimel |
| 12:00 | Lunch | |
| 1:00 | Introduction to the HDF5 File Format (download presentation PDF) | Lindsay Powers |
| 1:45 | An Introduction to Hyperspectral Remote Sensing (video) | Tristan Goulden |
| 2:00 | Work with Hyperspectral Remote Sensing data in R - HDF5 | Leah & Naupaka |
| 2:45 | NEON Hyperspectral Remote Sensing Data in R - Efficient Processing Using Functions | Leah & Naupaka |
| 3:15 | Plot a Spectral Signature from Hyperspectral Remote Sensing data in R - HDF5 | Leah & Naupaka |
| 3:45 | Break | |
| 4:00 | Calculate NDVI from NEON Hyperspectral Remote Sensing Data in R | Leah & Naupaka |
| 6:00 | Dinner Break | |
| 8:00 | Reproducible Science Methods | Naupaka |
| 10:00 | End |
Additional Resources
These tutorials were not part of the day's curriculum but may be useful to those looking for more background information on raster data and HDF5 formats or to go beyond the day's materials
- Introduction to the HDF5 File Format - Using HDFView & R Tutorial Series
- Raster Graphics Supplemental
- Subset HDF5 file in R
- Extract Spectra using Masks in R
Tuesday: LiDAR & Automation
In the morning, we will review the basics of discrete return and full waveform lidar data. In the afternoon, we will focus on automation as a means to write more efficient, usable code.
After completing these activities, you will be able to:
- Explain the difference between active and passive sensors.
- Explain the difference between discrete return and full waveform LiDAR.
- Describe applications of LiDAR remote sensing data in the natural sciences.
- Describe several NEON LiDAR remote sensing data products.
- Explain why modularization is important and supports efficient coding practices.
- How to modularize code using functions.
- Integrate basic automation into your existing data workflow.
| Time | Topic | Instructor |
|---|---|---|
| 8:00 | Introduction to LiDAR (video) | Tristan Goulden |
| 8:30 | Introduction to full waveform LiDAR (video) | Keith Krause |
| 9:00 | OpenTopography: Increasing the Impact of High Resolution Topography through Open, Online Access to Data and Processing (download presentation PDF) | Christopher Crosby |
| 10:00 | Break | |
| 10:15 | Classify a Raster using Threshold Values in R | Leah, Naupaka |
| 11:00 | Mask a Raster using Threshold Values in R | Leah, Naupaka |
| 12:00 | Lunch | |
| 1:00 | Code Automation - Adapted from Reproducible Science Curriculum materials | Naupaka |
| 5:30 | End |
Additional Resources
These tutorials were not part of the day's curriculum but may be useful to those looking for more background information on raster data or to go beyond the day's materials
- Primer on Raster Data in R tutorial series
- Introduction to Working with Raster Data in R tutorial series
- Overlay function in Raster Calculations tutorial: Use the overlay function to perform efficient raster processing tasks
- Create A Hillshade from a Terrain Raster in R
- Dealing with Spatial Extents when working with Heterogeneous Data
Wednesday: Comparing Ground to Airborne – Uncertainty
Today, we will focus on the importance of uncertainty when using remote sensing data. We will work on a hands-on activity where we compare remote sensing derived vegetation metrics to metrics collected on the ground (in situ).
- Explain sources of uncertainty in remote sensing data.
- Measure the differences between a metric derived from remote sensing data and the same metric derived from data collected on the ground.
| Time | Topic | Instructor |
|---|---|---|
| 8:00 | Vegetation Data Indices and NEON Data Products (video) | Dave Hulslander |
| 8:45 | NEON Terrestrial Observation Vegetation Sampling & Integration with Remote Sensing (video) | Courtney Meier |
| 9:30 | The Importance of Validation & Uncertainty Issues when Using Remote Sensing Data | Kyla Dahlin |
| 10:00 | Break | |
| 10:15 | Extract Values from Rasters in R & Compare Ground to Airborne | Kyla |
| 12:00 | Lunch | |
| 1:00 | Collaborative mini-projects in Uncertainty | Leah & Kyla |
| 3:30 | Break | |
| 3:45 | Collaborative mini-projects in Uncertainty, continued | |
| 5:00 | Present Results & Methods Discussion | |
| 6:00 | End |
Additional Resources
These three presentations (videos linked) were not part of the 2016 Data Institute but the topics are aligned with the day's theme and may be of interest.
Thursday: LiDAR & Hyperspectral Data Fusion
Today, we will focus on the importance of uncertainty when using remote sensing data. We will work on a hands-on activity where we compare remote sensing derived vegetation metrics to metrics collected on the ground (in situ).
- Use the thresholding approach to data fusion that masks and identify areas of a site with similar physical and other characteristics (e.g., the same slope, aspect, elevation, vegetation height).
- Describe a statistical approach to data fusion.
| Time | Topic | Instructor |
|---|---|---|
| 8:00 | Combining LiDAR & Hyperspectral Data to Advance Ecology | Kyla Dahlin |
| 9:00 | Thresholding | Kyla |
| 10:15 | Break | |
| 10:30 | Data Fusion Group Coding Activity | Kyla |
| 12:00 | Lunch | |
| 1:00 | Data Fusion Group Coding Activity | Kyla |
| 3:30 | Break | |
| 3:45 | NEON facilities tour | |
| 5:15 | Group Project Selection & Prep | |
| 6:00 | End |
Friday: Data Institute Capstone Projects
Today, you will use all of the skills you’ve learned at the Institute to work on a team project that uses NEON and/or related data!
- Apply the skills that you have learned to process data using efficient coding practices.
- Expand upon your skills in working with remote sensing data through collaborative peer-learning.
- Apply your understanding of remote sensing data and use it to address a science question of your choice
- Implement version control and collaborate with your colleagues through the GitHub platform.
Throughout the day teams will be working on their capstone projects. Data Institute instructors and other NEON project scientists will be available to answer questions and assist as needed with the projects.
| Time | Topic | Instructor |
|---|---|---|
| 9:00 | Breakout rooms open for teams to work on capstone project | |
| 12:00 | Lunch | |
| 1:00 | Breakout rooms open for teams to work | |
| 4:45 | End of Day Wrap Up & Presentation Sign Ups | |
| 6:30 | Participants must leave building for night |
Additional Resources
If you are interested in creating a reveal.js presentation from R Markdown to present your capstone project, you can find out more information here: RStudio's reveal.js documentation.
Saturday: Data Institute Capstone Project Presentations
| Time | Topic | Instructor |
|---|---|---|
| 9:00 | Presentations Start | |
| 12:00 | Lunch | |
| 1:00 | Final Questions & Institute Debrief | |
| 2:00 | End |
Communal Notetaking
During the Data Institute we will use Etherpad as a source for communal notetaking. 2016 Data Institute Etherpad