Workshop
NEON Data Institute 2017: Remote Sensing with Reproducible Workflows using Python
NEON
-
NEON Data Institutes provide critical skills and foundational knowledge for graduate students and early career scientists working with heterogeneous spatio-temporal data to address ecological questions.
Data Institute Overview
The 2017 Institute focuses on remote sensing of vegetation using open source tools and reproducible science workflows -- the primary programming language will be Python.
This Institute will be held at NEON headquarters in June 2017. In addition to the six days of in-person training, there are three weeks of pre-institute materials is to ensure that everyone comes to the Institute ready to work in a collaborative research environment. Pre-institute materials are online & individually paced, expect to spend 1-5 hrs/week depending on your familiarity with the topic.
Schedule
| Time | Day | Description |
|---|---|---|
| -- | Computer Setup Materials | |
| -- | 25 May - 1 June | Intro to NEON & Reproducible Science |
| -- | 2-8 June | Version Control & Collaborative Science with Git & GitHub |
| -- | 9-15 June | Documentation of Your Workflow with iPython/Jupyter Notebooks |
| -- | 19-24 June | Data Institute |
| 7:50am - 6:30 pm | Monday | Intro to NEON, Intro to HDF5 & Hyperspectral Remote Sensing |
| 8:00am - 6:30pm | Tuesday | Reproducible & Automated Workflows, Intro to LiDAR data |
| 8:00am - 6:30pm | Wednesday | Remote Sensing Uncertainty |
| 8:00am - 6:30pm | Thursday | Hyperspectral Data & Vegetation |
| 8:00am - 6:30pm | Friday | Individual/Group Applications |
| 9:00am - 1:00pm | Saturday | Group Application Presentations |
Key 2017 Dates
- Applications Open: 17 January 2017
- Application Deadline: 10 March 2017
- Notification of Acceptance: late March 2017
- Tuition payment due by: mid April 2017
- Pre-institute online activities: June 1-17, 2017
- Institute Dates: June 19-24, 2017
Instructors
Dr. Tristan Goulden, Associate Scientist-Airborne Platform, Battelle-NEON: Tristan is a remote sensing scientist with NEON specializing in LiDAR. He also co-lead NEON’s Remote Sensing IPT (integrated product team) which focusses on developing algorithms and associated documentation for all of NEON’s remote sensing data products. His past research focus has been on characterizing uncertainty in LiDAR observations/processing and propagating the uncertainty into downstream data products. During his PhD, he focused on developing uncertainty models for topographic attributes (elevation, slope, aspect), hydrological products such as watershed boundaries, stream networks, as well as stream flow and erosion at the watershed scale. His past experience in LiDAR has included all aspects of the LIDAR workflow including; mission planning, airborne operations, processing of raw data, and development of higher level data products. During his graduate research he applied these skills on LiDAR flights over several case study watersheds of study as well as some amazing LiDAR flights over the Canadian Rockies for monitoring change of alpine glaciers. His software experience for LiDAR processing includes Applanix’s POSPac MMS, Optech’s LMS software, Riegl’s LMS software, LAStools, Pulsetools, TerraScan, QT Modeler, ArcGIS, QGIS, Surfer, and self-written scripts in Matlab for point-cloud, raster, and waveform processing.
Bridget Hass, Remote Sensing Data Processing Technician, Battelle-NEON: Bridget’s daily work includes processing LiDAR and hyperspectral data collected by NEON's Aerial Observation Platform (AOP). Prior to joining NEON, Bridget worked in marine geophysics as a shipboard technician and research assistant. She is excited to be a part of producing NEON's AOP data and to share techniques for working with this data during the 2017 Data Institute.
Dr. Naupaka Zimmerman, Assistant Professor of Biology, University of San Francisco: Naupaka’s research focuses on the microbial ecology of plant-fungal interactions. Naupaka brings to the course experience and enthusiasm for reproducible workflows developed after discovering how challenging it is to keep track of complex analyses in his own dissertation and postdoctoral work. As a co-founder of the International Network of Next-Generation Ecologists and an instructor and lesson maintainer for Software Carpentry and Data Carpentry, Naupaka is very interested in providing and improving training experiences in open science and reproducible research methods.
Dr. Paul Gader, Professor, University of Florida:
Paul is a Professor of Computer & Information Science & Engineering (CISE)
at the Engineering School of Sustainable Infrastructure and the Environment
(ESSIE) at the University of Florida(UF). Paul received his Ph.D. in Mathematics
for parallel image processing and applied mathematics research in 1986 from UF,
spent 5 years in industry, and has been teaching at various universities since 1991.
His first research in image processing was in 1984 focused on algorithms
for detection of bridges in Forward Looking Infra-Red (FLIR) imagery. He has
investigated algorithms for land mine research since 1996, leading a team that
produced new algorithms and real-time software for a sensor system currently
operational in Afghanistan. His landmine detection projects involve algorithm
development for data generated from hand-held, vehicle-based, and airborne
sensors, including ground penetrating radar, acoustic/seismic, broadband IR
(emissive and reflective bands), emissive and reflective hyperspectral imagery,
and wide-band electro-magnetic sensors. In the past few years, he focused on
algorithms for imaging spectroscopy. He is currently researching nonlinear
unmixing for object and material detection, classification and segmentation, and
estimating plant traits. He has given tutorials on nonlinear unmixing at
International Conferences. He is a Fellow of the Institute of Electrical and
Electronic Engineers, an Endowed Professor at the University of Florida, was
selected for a 3-year term as a UF Research Foundation Professor, and has over
100 refereed journal articles and over 300 conference articles.
This page includes all of the materials needed for the Data Institute including the pre-institute materials. Please use the sidebar menu to find the appropriate week or day. If you have problems with any of the materials please email us or use the comments section at the bottom of the appropriate page.
Pre-Institute: Computer Set Up Materials
It is important that you have your computer setup, prior to diving into the pre-institute materials in week 2! Please review the links below to setup the laptop you will be bringing to the Data Institute.
Let's Get Your Computer Setup!
Go to each of the following tutorials and complete the directions to set your computer up for the Data Institute.
Install Git, Bash Shell, Python
This page outlines the tools and resources that you will need to install Git, Bash and Python applications onto your computer as the first step of our Python skills tutorial series.
Checklist
Detailed directions to accomplish each objective are below.
- Install Bash shell (or shell of preference)
- Install Git
- Install Python 3.x
Bash/Shell Setup
Install Bash for Windows
- Download the Git for Windows installer.
- Run the installer and follow the steps bellow:
- Welcome to the Git Setup Wizard: Click on "Next".
- Information: Click on "Next".
- Select Destination Location: Click on "Next".
- Select Components: Click on "Next".
- Select Start Menu Folder: Click on "Next".
- Adjusting your PATH environment: Select "Use Git from the Windows Command Prompt" and click on "Next". If you forgot to do this programs that you need for the event will not work properly. If this happens rerun the installer and select the appropriate option.
- Configuring the line ending conversions: Click on "Next". Keep "Checkout Windows-style, commit Unix-style line endings" selected.
- Configuring the terminal emulator to use with Git Bash: Select "Use Windows' default console window" and click on "Next".
- Configuring experimental performance tweaks: Click on "Next".
- Completing the Git Setup Wizard: Click on "Finish".
This will provide you with both Git and Bash in the Git Bash program.
Install Bash for Mac OS X
The default shell in all versions of Mac OS X is bash, so no
need to install anything. You access bash from the Terminal
(found in
/Applications/Utilities). You may want to keep
Terminal in your dock for this workshop.
Install Bash for Linux
The default shell is usually Bash, but if your
machine is set up differently you can run it by opening a
terminal and typing bash. There is no need to
install anything.
Git Setup
Git is a version control system that lets you track who made changes to what when and has options for easily updating a shared or public version of your code on GitHub. You will need a supported web browser (current versions of Chrome, Firefox or Safari, or Internet Explorer version 9 or above).
Git installation instructions borrowed and modified from Software Carpentry.
Git for Windows
Git should be installed on your computer as part of your Bash install.Git on Mac OS X
Video Tutorial
Install Git on Macs by downloading and running the most recent installer for
"mavericks" if you are using OS X 10.9 and higher -or- if using an
earlier OS X, choose the most recent "snow leopard" installer, from
this list.
After installing Git, there will not be anything in your
/Applications folder, as Git is a command line program.
Git on Linux
If Git is not already available on your machine you can try to
install it via your distro's package manager. For Debian/Ubuntu run
sudo apt-get install git and for Fedora run
sudo yum install git.
Setting Up Python
Python is a popular language for scientific computing and data science, as well as being a great for general-purpose programming. Installing all of the scientific packages individually can be a bit difficult, so we recommend using an all-in-one installer, like Anaconda.
Regardless of how you choose to install it, **please make sure your environment
is set up with Python version 3.7 (at the time of writing, the gdal package did not work
with the newest Python version 3.6). Python 2.x is quite different from Python 3.x
so you do need to install 3.x and set up with the 3.7 environment.
We will teach using Python in the Jupyter Notebook environment, a programming environment that runs in a web browser. For this to work you will need a reasonably up-to-date browser. The current versions of the Chrome, Safari and Firefox browsers are all supported (some older browsers, including Internet Explorer version 9 and below, are not). You can choose to not use notebooks in the course, however, we do recommend you download and install the library so that you can explore this tool.
Windows
Download and install Anaconda. Download the default Python 3 installer (3.7). Use all of the defaults for installation except make sure to check Make Anaconda the default Python.
Mac OS X
Download and install Anaconda. Download the Python 3.x installer, choosing either the graphical installer or the command-line installer (3.7). For the graphical installer, use all of the defaults for installation. For the command-line installer open Terminal, navigate to the directory with the download then enter:
bash Anaconda3-2020.11-MacOSX-x86_64.sh (or whatever you file name is)
Linux
Download and install Anaconda. Download the installer that matches your operating system and save it in your home folder. Download the default Python 3 installer.Open a terminal window and navigate to your downloads folder. Type
bash Anaconda3-2020.11-Linux-ppc64le.sh
and then press tab. The name of the file you just downloaded should appear.
Press enter. You will follow the text-only prompts. When there is a colon at
the bottom of the screen press the down arrow to move down through the text.
Type yes and press enter to approve the license. Press enter to
approve the default location for the files. Type yes and press
enter to prepend Anaconda to your PATH (this makes the Anaconda
distribution the default Python).
Install Python packages
We need to install several packages to the Python environment to be able to work with the remote sensing data
- gdal
- h5py
If you are new to working with command line you may wish to complete the next setup instructions which provides and intro to command line (bash) prior to completing these package installation instructions.
Windows
Create a new Python 3.7 environment by opening Windows Command Prompt and typing
conda create –n py37 python=3.7 anaconda
When prompted, activate the py37 environment in Command Prompt by typing
activate py37
You should see (py37) at the beginning of the command line. You can also test
that you are using the correct version by typing python --version.
Install Python package(s):
- gdal:
conda install gdal - h5py:
conda install h5py
Note: You may need to only install gdal as the others may be included in the default.
Mac OS X
Create a new Python 3.7 environment by opening Terminal and typing
conda create –n py37 python=3.7 anaconda
This may take a minute or two.
When prompted, activate the py37 environment in Command Prompt by typing
source activate py37
You should see (py37) at the beginning of the command line. You can also test
that you are using the correct version by typing python --version.
Install Python package(s):
- gdal:
conda install gdal - h5py:
conda install h5py
Linux
Open default terminal application (on Ubuntu that will be gnome-terminal).Launch Python.
Install Python package(s):
- gdal:
conda install gdal - h5py:
conda install h5py
Set up Jupyter Notebook Environment
In your terminal application, navigate to the directory (cd) that where you
want the Jupyter Notebooks to be saved (or where they already exist).
Open Jupyter Notebook with
jupyter notebook
Once the notebook is open, check which version of Python you are in by using the prompts
# check what version of Python you are using.
import sys
sys.version
You should now be able to work in the notebook.
The gdal package that occasionally has problems with some versions of Python.
Therefore test out loading it using
import gdal.
Additional Resources
- Setting up the Python Environment section from the Python Bootcamp
- Conda Help: setting up an environment
- iPython documentation: Kernals
Set up GitHub Working Directory - Quick Intro to Bash
Checklist
Once you have Git and Bash installed, you are ready to configure Git.
On this page you will:
- Create a directory for all future GitHub repositories created on your computer
To ensure Git is properly installed and to create a working directory for GitHub, you will need to know a bit of shell -- brief crash course below.
Crash Course on Shell
The Unix shell has been around longer than most of its users have been alive. It has survived so long because it’s a power tool that allows people to do complex things with just a few keystrokes. More importantly, it helps them combine existing programs in new ways and automate repetitive tasks so they aren’t typing the same things over and over again. Use of the shell is fundamental to using a wide range of other powerful tools and computing resources (including “high-performance computing” supercomputers).
This section is an abbreviated form of Software Carpentry’s The Unix Shell for Novice’s workshop lesson series. Content and wording (including all the above) is heavily copied and credit is due to those creators (full author list).
Our goal with shell is to:
- Set up the directory where we will store all of the GitHub repositories during the Institute,
- Make sure Git is installed correctly, and
- Gain comfort using bash so that we can use it to work with Git & GitHub.
Accessing Shell
How one accesses the shell depends on the operating system being used.
- OS X: The bash program is called Terminal. You can search for it in Spotlight.
- Windows: Git Bash came with your download of Git for Windows. Search Git Bash.
- Linux: Default is usually bash, if not, type
bashin the terminal.
Bash Commands
$
The dollar sign is a prompt, which shows us that the shell is waiting for input; your shell may use a different character as a prompt and may add information before the prompt.
When typing commands, either from these tutorials or from other sources, do not
type the prompt ($), only the commands that follow it.
In these tutorials, subsequent lines that follow a prompt and do not start with
$ are the output of the command.
listing contents - ls
Next, let's find out where we are by running a command called pwd -- print
working directory. At any moment, our current working directory is our
current default directory. I.e., the directory that the computer assumes we
want to run commands in unless we explicitly specify something else. Here, the
computer's response is /Users/neon, which is NEON’s home directory:
$ pwd
/Users/neon
If you are not, by default, in your home directory, you get there by typing:
$ cd ~
Now let's learn the command that will let us see the contents of our own
file system. We can see what's in our home directory by running ls --listing.
$ ls
Applications Documents Library Music Public
Desktop Downloads Movies Pictures
(Again, your results may be slightly different depending on your operating system and how you have customized your filesystem.)
ls prints the names of the files and directories in the current directory in
alphabetical order, arranged neatly into columns.
Change directory -- cd
Now we want to move into our Documents directory where we will create a
directory to host our GitHub repository (to be created in Week 2). The command
to change locations is cd followed by a directory name if it is a
sub-directory in our current working directory or a file path if not.
cd stands for "change directory", which is a bit misleading: the command
doesn't change the directory, it changes the shell's idea of what directory we
are in.
To move to the Documents directory, we can use the following series of commands to get there:
$ cd Documents
These commands will move us from our home directory into our Documents
directory. cd doesn't print anything, but if we run pwd after it, we can
see that we are now in /Users/neon/Documents.
If we run ls now, it lists the contents of /Users/neon/Documents, because
that's where we now are:
$ pwd
/Users/neon/Documents
$ ls
data/ elements/ animals.txt planets.txt sunspot.txt
To use cd, you need to be familiar with paths, if not, read the section on
Full, Base, and Relative Paths .
Make a directory -- mkdir
Now we can create a new directory called GitHub that will contain our GitHub
repositories when we create them later.
We can use the command mkdir NAME-- “make directory”
$ mkdir GitHub
There is not output.
Since GitHub is a relative path (i.e., doesn't have a leading slash), the
new directory is created in the current working directory:
$ ls
data/ elements/ GitHub/ animals.txt planets.txt sunspot.txt
Is Git Installed Correctly?
All of the above commands are bash commands, not Git specific commands. We still need to check to make sure git installed correctly. One of the easiest ways is to check to see which version of git we have installed.
Git commands start with git.
We can use git --version to see which version of Git is installed
$ git --version
git version 2.5.4 (Apple Git-61)
If you get a git version number, then Git is installed!
If you get an error, Git isn’t installed correctly. Reinstall and repeat.
Setup Git Global Configurations
Now that we know Git is correctly installed, we can get it set up to work with.
The text below is modified slightly from Software Carpentry's Setting up Git lesson.
When we use Git on a new computer for the first time, we need to configure a few things. Below are a few examples of configurations we will set as we get started with Git:
- our name and email address,
- to colorize our output,
- what our preferred text editor is,
- and that we want to use these settings globally (i.e. for every project)
On a command line, Git commands are written as git verb, where verb is what
we actually want to do.
Set up you own git with the following command, using your own information instead of NEON's.
$ git config --global user.name "NEON Science"
$ git config --global user.email "neon@BattelleEcology.org"
$ git config --global color.ui "auto"
Then set up your favorite text editor following this table:
| Editor | Configuration command |
|---|---|
| nano | $ git config --global core.editor "nano -w" |
| Text Wrangler | $ git config --global core.editor "edit -w" |
| Sublime Text (Mac) | $ git config --global core.editor "subl -n -w" |
| Sublime Text (Win, 32-bit install) | $ git config --global core.editor "'c:/program files (x86)/sublime text 3/sublime_text.exe' -w" |
| Sublime Text (Win, 64-bit install) | $ git config --global core.editor "'c:/program files/sublime text 3/sublime_text.exe' -w" |
| Notepad++ (Win) | $ git config --global core.editor "'c:/program files (x86)/Notepad++/notepad++.exe' -multiInst -notabbar -nosession -noPlugin" |
| Kate (Linux) | $ git config --global core.editor "kate" |
| Gedit (Linux) | $ git config --global core.editor "gedit -s -w" |
| emacs | $ git config --global core.editor "emacs" |
| vim | $ git config --global core.editor "vim" |
The four commands we just ran above only need to be run once:
the flag --global tells Git to use the settings for every project in your user
account on this computer.
You can check your settings at any time:
$ git config --list
You can change your configuration as many times as you want; just use the same commands to choose another editor or update your email address.
Now that Git is set up, you will be ready to start the Week 2 materials to learn about version control and how Git & GitHub work.
Install QGIS & HDF5View
Install HDFView
The free HDFView application allows you to explore the contents of an HDF5 file.
To install HDFView:
-
Click to go to the download page.
-
From the section titled HDF-Java 2.1x Pre-Built Binary Distributions select the HDFView download option that matches the operating system and computer setup (32 bit vs 64 bit) that you have. The download will start automatically.
-
Open the downloaded file.
- Mac - You may want to add the HDFView application to your Applications directory.
- Windows - Unzip the file, open the folder, run the .exe file, and follow directions to complete installation.
- Open HDFView to ensure that the program installed correctly.
Install QGIS
QGIS is a free, open-source GIS program. Installation is optional for the 2018 Data Institute. We will not directly be working with QGIS, however, some past participants have found it useful to have during the capstone projects.
To install QGIS:
Download the QGIS installer on the QGIS download page here. Follow the installation directions below for your operating system.
Windows
- Select the appropriate QGIS Standalone Installer Version for your computer.
- The download will automatically start.
- Open the .exe file and follow prompts to install (installation may take a while).
- Open QGIS to ensure that it is properly downloaded and installed.
Mac OS X
- Select KyngChaos QGIS download page. This will take you to a new page.
- Select the current version of QGIS. The file download (.dmg format) should start automatically.
- Once downloaded, run the .dmg file. When you run the .dmg, it will create a directory of installer packages that you need to run in a particular order. IMPORTANT: read the READ ME BEFORE INSTALLING.rtf file!
Install the packages in the directory in the order indicated.
- GDAL Complete.pkg
- NumPy.pkg
- matplotlib.pkg
- QGIS.pkg - NOTE: you need to install GDAL, NumPy and matplotlib in order to successfully install QGIS on your Mac!
Once all of the packages are installed, open QGIS to ensure that it is properly installed.
LINUX
- Select the appropriate download for your computer system.
- Note: if you have previous versions of QGIS installed on your system, you may run into problems. Check out
Pre-Institute Week 1: NEON & Reproducible Science
In the first week of the pre-institute activities, we will review the NEON project. We will also provide you with a general overview of reproducible science. Over the next few weeks will we ask you to review materials and submit something that demonstrates you have mastered the materials.
Learning Objectives
After completing these activities, you will be able to:
- Describe the NEON project, the data collected, and where to access more information about the project.
- Know how to access other code resources for working with NEON data.
- Explain why reproducible workflows are useful and important in your research.
Week 1 Assignment
After reviewing the materials below, please write up a summary of a project that you are interested working on at the Data Institute. Be sure to consider what data you will need (NEON or other). You will have time to refine your idea over the next few weeks. Save this document as you will submit it next week as a part of week 2 materials!
Deadline: Please complete this by Thursday June 1st @ 11:59 MDT.
Week 1 Materials
Please carefully read and review the materials below:
Introduction to the National Ecological Observatory Network (NEON)
Here we will provide an overview of the National Ecological Observatory Network (NEON). Please carefully read through these materials and links that discuss NEON’s mission and design.
Learning Objectives
At the end of this activity, you will be able to:
- Explain the mission of the National Ecological Observatory Network (NEON).
- Explain the how sites are located within the NEON project design.
- Explain the different types of data that will be collected and provided by NEON.
The NEON Project Mission & Design
To capture ecological heterogeneity across the United States, NEON’s design divides the continent into 20 statistically different eco-climatic domains. Each NEON field site is located within an eco-climatic domain.
The Science and Design of NEON
To gain a better understanding of the broad scope fo NEON watch this 4 minute long video.
Please, read the following page about NEON's mission.
Data Institute Participants -- Thought Question: How might/does the NEON project intersect with your current research or future career goals?
NEON's Spatial Design
The Spatial Design of NEON
Watch this 4:22 minute video exploring the spatial design of NEON field sites.
Please read the following page about NEON's Spatial Design:
Read this primer on NEON's Sampling Design
Read about the different types of field sites - core and relocatable
NEON Field Site Locations
Explore the NEON Field Site map taking note of the locations of
- Aquatic & terrestrial field sites.
- Core & relocatable field sites.
Explore the NEON field site map. Do the following:
- Zoom in on a study area of interest to see if there are any NEON field sites that are nearby.
- Use the menu below the map to filter sites by name, type, domain, or state.
- Select one field site of interest.
- Click on the marker in the map.
- Then click on Site Details to jump to the field site landing page.
Data Institute Participant -- Thought Questions: Use the map above to answer these questions. Consider the research question that you may explore as your Capstone Project at the Institute or about a current project that you are working on and answer the following questions:
- Are there NEON field sites that are in study regions of interest to you?
- What domains are the sites located in?
- What NEON field sites do your current research or Capstone Project ideas coincide with?
- Is the site(s) core or relocatable?
- Is it/are they terrestrial or aquatic?
- Are there data available for the NEON field site(s) that you are most interested in? What kind of data are available?
Data Tip: You can download maps, kmz, or shapefiles of the field sites here.
NEON Data
How NEON Collects Data
Watch this 3:06 minute video exploring the data that NEON collects.
Read the Data Collection Methods page to learn more about the different types of data that NEON collects and provides. Then, follow the links below to learn more about each collection method:
- Aquatic Observation System (AOS)
- Aquatic Instrument System (AIS)
- Terrestrial Instrument System (TIS) -- Flux Tower
- Terrestrial Instrument System (TIS) -- Soil Sensors and Measurements
- Terrestrial Organismal System (TOS)
- Airborne Observation Platform (AOP)
All data collection protocols and processing documents are publicly available. Read more about the standardized protocols and how to access these documents.
Specimens & Samples
NEON also collects samples and specimens from which the other data products are based. These samples are also available for research and education purposes. Learn more: NEON Biorepository.
Airborne Remote Sensing
Watch this 5 minute video to better understand the NEON Airborne Observation Platform (AOP).
Data Institute Participant – Thought Questions: Consider either your current or future research or the question you’d like to address at the Institute.
- Which types of NEON data may be more useful to address these questions?
- What non-NEON data resources could be combined with NEON data to help address your question?
- What challenges, if any, could you foresee when beginning to work with these data?
Data Tip: NEON also provides support to your own research including proposals to fly the AOP over other study sites, a mobile tower/instrumentation setup and others. Learn more here the Assignable Assets programs .
Access NEON Data
NEON data are processed and go through quality assurance quality control checks at NEON headquarters in Boulder, CO. NEON carefully documents every aspect of sampling design, data collection, processing and delivery. This documentation is freely available through the NEON data portal.
- Visit the NEON Data Portal - data.neonscience.org
- Read more about the quality assurance and quality control processes for NEON data and how the data are processed from raw data to higher level data products.
- Explore NEON Data Products. On the page for each data product in the catalog you can find the basic information about the product, find the data collection and processing protocols, and link directly to downloading the data.
- Additionally, some types of NEON data are also available through the data portals of other organizations. For example, NEON Terrestrial Insect DNA Barcoding Data is available through the Barcode of Life Datasystem (BOLD). Or NEON phenocam images are available from the Phenocam network site. More details on where else the data are available from can be found in the Availability and Download section on the Product Details page for each data product (visit Explore Data Products to access individual Product Details pages).
Pathways to access NEON Data
There are several ways to access data from NEON:
- Via the NEON data portal. Explore and download data. Note that much of the tabular data is available in zipped .csv files for each month and site of interest. To combine these files, use the neonUtilities package (R tutorial, Python tutorial).
- Use R or Python to programmatically access the data. NEON and community members have created code packages to directly access the data through an API. Learn more about the available resources by reading the Code Resources page or visiting the NEONScience GitHub repo.
- Using the NEON API. Access NEON data directly using a custom API call.
- Access NEON data through partner's portals. Where NEON data directly overlap with other community resources, NEON data can be accessed through the portals. Examples include Phenocam, BOLD, Ameriflux, and others. You can learn more in the documentation for individual data products.
Data Institute Participant – Thought Questions: Use the Data Portal tools to investigate the data availability for the field sites you’ve already identified in the previous Thought Questions.
- What types of aquatic/terrestrial data are currently available? Remote sensing data?
- Of these, what type of data are you most interested in working with for your project while at the Institute.
- For what time period does the data cover?
- What format is the downloadable file available in?
- Where is the metadata to support this data?
Data Institute Participants: Intro to NEON Culmination Activity
Write up a brief summary of a project that you might want to explore while at the Data Institute in Boulder, CO. Include the types of NEON (and other data) that you will need to implement this project. Save this summary as you will be refining and adding to your ideas over the next few weeks.
The goal of this activity if for you to begin to think about a Capstone Project that you wish to work on at the end of the Data Institute. This project will ideally be performed in groups, so over the next few weeks you'll have a chance to view the other project proposals and merge projects to collaborate with your colleagues.
The Importance of Reproducible Science
Verifiability and reproducibility are among the cornerstones of the scientific process. They are what allows scientists to "stand on the shoulder of giants". Maintaining reproducibility requires that all data management, analysis, and visualization steps behind the results presented in a paper are documented and available in full detail. Reproducibility here means that someone else should either be able to obtain the same results given all the documented inputs and the published instructions for processing them, or if not, the reasons why should be apparent. From Reproducible Science Curriculum
- Summarize the four facets of reproducibility.
- Describe several ways that reproducible workflows can improve your workflow and research.
- Explain several ways you can incorporate reproducible science techniques into your own research.
Getting Started with Reproducible Science
Please view the online slide-show below which summarizes concepts taught in the Reproducible Science Curriculum.
View Reproducible Science Slideshow
A Gap In Understanding

Reproducibility and Your Research
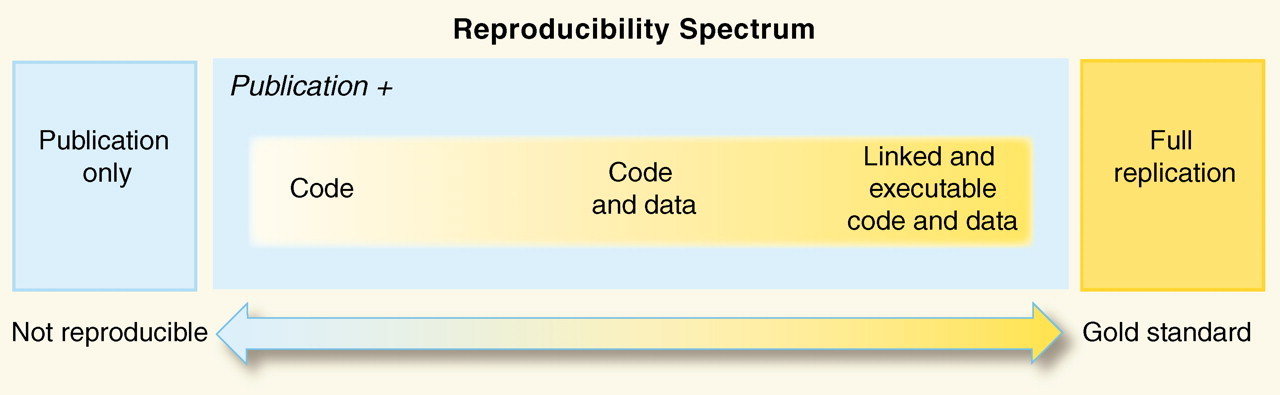
How reproducible is your current research?
View Reproducible Science Checklist
- Do you currently apply any of the items in the checklist to your research?
- Are there elements in the list that you are interested in incorporating into your workflow? If so, which ones?
Additional Readings (optional)
- Nature has collated and published (with open-access) a special archive on the Challenges of Irreproducible Science .
- The Nature Publishing group has also created a Reporting Checklist for its authors that focuses primaily on reporting issues but also includes sections for sharing code.
- Recent open-access issue of Ecography focusing on reproducible ecology and software packages available for use.
- A nice short blog post with an annotated bibliography of "Top 10 papers discussing reproducible research in computational science" from Lorena Barba: Barba group reproducibility syllabus.
Pre-Institute Week 2: Version Control & Collaborative Science
The goal of the pre-institute materials is to ensure that everyone comes to the Institute ready to work in a collaborative research environment. If you recall, from last week, the four facets of reproducibility are documentation, organization, automation, and dissemination.
This week we will focus on learning to use tools to help us with these facets: Git and GitHub. The Git Hub environment supports both a collaborative approach to science through code sharing and dissemination, and a powerful version control system that supports both efficient project organization, and an effective way to save your work.
Learning Objectives
After completing these activities, you will be able to:
- Summarize the key components of a version control system
- Know how to setup a GitHub account
- Know how to setup Git locally
- Work in a collaborative workflow on GitHub
Week 2 Assignment
The assignment for this week is to revise the Data Institute capstone project summary that you developed last week. You will submit your project summary, with a brief biography to introduce yourself, to a shared GitHub repository.
Please complete this assignment by Thursday June 8th @ 11:59 PM MDT.
If you are familiar with forked repos and pull requests GitHub, and the use of Git in the command line, you may be able to complete the assignment without viewing the
tutorials.
Assignment: Version Control with GitHub
DUE: 21 June 2018
During the NEON Data Institute, you will share the code that you create daily
with everyone on the NEONScience/DI-NEON-participants repo.
Through this week’s tutorials, you have learned the basic skills needed to successfully share your work at the Institute including how to:
- Create your own GitHub user account,
- Set up Git on your computer (please do this on the computer you will be bringing to the Institute), and
- Create a Markdown file with a biography of yourself and the project you are interested in working on at the Institute. This biography was shared with the group via the Data Institute’s GitHub repo.
Checklist for this week’s Assignment:
You should have completed the following after Pre-institute week 2:
- Fork & clone the NEON-DataSkills/DI-NEON-participants repo.
- Create a .md file in the
participants/2018-RemoteSensing/pre-institute2-gitdirectory of the repo. Name the document LastName-FirstName.md. - Write a biography that introduces yourself to the other participants. Please
provide basic information including:
- name,
- domain of interest,
- one goal for the course,
- an updated version of your Capstone Project idea,
- and the list of data (NEON or other) to support the project that you created during last week’s materials.
- Push the document from your local computer to your GithHub repo.
- Created a Pull Request to merge this document back into the NEON-DataSkills/DI-NEON-participants repo.
NOTE: The Data Institute repository is a public repository, so all members of the Institute, as well as anyone in the general public who stumbles on the repo, can see the information. If you prefer not to share this information publicly, please submit the same document but use a pseudonym (cartoon character names would work well) and email us with the pseudonym so that we can connect the submitted document to you.
Have questions? No problem. Leave your question in the comment box below. It's likely some of your colleagues have the same question, too! And also likely someone else knows the answer.
Week 2 Materials
Please complete each of the short tutorials in this series.
Version Control with GitHub
Pre-Institute Week 3: Documentation of Your Workflow
In week 3, you will use Jupyter Notebooks (formerly iPython Notebooks) to document code and efficiently publish code results & outputs. You will practice your Git skills by publishing your work in the NEON-WorkWithData/DI-NEON-participants GitHub repository.
In addition, you will watch a video that provides an overview of the NEON Vegitation Indices that are available as data products in preparation for Monday's materials.
Learning Objectives
After completing these activities, you will be able to:
- Use Jupyter Notebooks to create code with formatted context text
- Describe the value of documented workflows
Week 3 Assignment, Part 1
Please complete the activity and submit your work to the GitHub repo by Thursday June 17th at 11:59 MDT.
If you are familiar with using Jupyter Notebooks to document your workflow and knitting to HTML then you may be able to complete the assignment without viewing the tutorials.
Assignment: Reproducible Workflows with Jupyter Notebooks
In this tutorial you will learn how to open a .tiff file in Jupyter Notebook and learn about kernels.
The goal of the activity is simply to ensure that you have basic
familiarity with Jupyter Notebooks and that the environment, especially the
gdal package is correctly set up before you pursue more programming tutorials. If you already
are familiar with Jupyter Notebooks using Python, you may be able to complete the
assignment without working through the instructions.
This will be accomplished by: *Create a new Jupyter kernel *Download a GEOTIFF file *Import file onto Jupyter Notebooks *Check the raster size
Assignment: Open a Tiff File in Jupyter Notebook
Set up Environment
First, we will set up the environment as you would need for each of the live coding sections of the Data Institute. The following directions are copied over from the Data Institute Set up Materials.
In your terminal application, navigate to the directory (cd) that where you
want the Jupyter Notebooks to be saved (or where they already exist).
We need to create a new Jupyter kernel for the Python 3.8 conda environment (py38) that Jupyter Notebooks will use.
In your Command Prompt/Terminal, type:
python -m ipykernel install --user --name py34 --display-name "Python 3.8 NEON-RSDI"
In your Command Prompt/Terminal, navigate to the directory (cd) that you
created last week in the GitHub materials. This is where the Jupyter Notebook
will be saved and the easiest way to access existing notebooks.
###Open Jupyter Notebook Open Jupyter Notebook by typing into a command terminal:
jupyter notebook
Once the notebook is open, check which version of Python you are in.
# Check what version of Python. Should be 3.8.
import sys
sys.version
To ensure that the correct kernel will operate, navigate to Kernel in the menu, select Kernel/Restart Kernel And Clear All Outputs.

You should now be able to work in the notebook.
#Download the digital terrain model (GEOTIFF file) Download the NEON GeoTiFF file of a digital terrain model (dtm) of the San Joaquin Experimental Range. Click this link to download dtm data: https://ndownloader.figshare.com/articles/2009586/versions/10. This will download a zippped full of data originally from a NEON data carpentry tutorial (https://datacarpentry.org/geospatial-workshop/data/).
Once downloaded, navigate through the folder to C:NEON-DS-Airborne-Remote-Sensing.zip\NEON-DS-Airborne-Remote-Sensing\SJER\DTM and save this file onto your own personal working directory. .
###Open GEOTIFF file in Jupyter Notebooks using gdal
The gdal package that occasionally has problems with some versions of Python.
Therefore test out loading it using:
import gdal.
If you have trouble, ensure that 'gdal' is installed on your current environment.
Establish your directory
Place the downloaded dtm file in a repository of your choice (or your current working directory). Navigate to that directory. wd= '/your-file-path-here' #Input the directory to where you saved the .tif file
Import the TIFF
Import the NEON GeoTiFF file of the digital terrain model (DTM)
from San Joaquin Experimental Range. Open the file using the gdal.Open command.Determine the size of the raster and (optional) plot the raster.
#Use GDAL to open GEOTIFF file stored in your directory SJER_DTM = gdal.Open(wd + 'SJER_dtmCrop.tif')>
#Determine the raster size.
SJER_DTM.RasterXSize
Add in both code chunks and text (markdown) chunks to fully explain what is done. If you would like to also plot the file, feel free to do so.
Push .ipynb to GitHub.
When finished, save as a .ipynb file.
Week 3 Materials
Please complete each of the short tutorials in this series.
Document Your Code with Jupyter Notebooks
Week 3 Assignment, Part 2
On the first day of the course, we will be working with hyperspectral data. Various indices, including the Normalized Difference Vegetation Index (NDVI), are common data products from hyperspectral data. In preparation for this content, please watch this video of David Hulslander discussing NEON remote sensing vegetation indices & data products.
Monday: HDF5, & Hyperspectral Data
Learning Objectives
After completing these activities, you will be able to:
- Open and work with raster data stored in HDF5 format in Python
- Explain the key components of the HDF5 data structure (groups, datasets and attributes)
- Open and use attribute data (metadata) from an HDF5 file in Python
Morning: Intro to NEON & HDF5
All activities are held in the the Classroom unless otherwise noted.
| Time | Topic | Instructor/Location |
|---|---|---|
| 8:00 | Welcome & Introductions | |
| 8:30 | Introduction to the National Ecological Observatory Network | Megan Jones |
| 9:00 | Introduction to NEON AOP (download presentation) | Nathan Leisso |
| 9:30 | NEON RGB Imagery (download presentation) | Bill Gallery |
| 10:00 | Introduction to the HDF5 File Format (download PDF) | Ted Haberman |
| 10:30 | BREAK | |
| 10:45 | NEON Tour | |
| 12:00 | LUNCH | Classroom/Patio |
Afternoon: Hyperspectral Remote Sensing
| Time | Topic | Instructor/Location |
|---|---|---|
| 13:00 | An Introduction to Hyperspectral Remote Sensing (related video) | Tristan Goulden |
| 13:20 | Work with Hyperspectral Remote Sensing data & HDF5 | |
| Explore NEON HDF5 format with Viewer | Tristan | |
| NEON AOP Hyperspectral Data in HDF5 format with Python | Bridget Hass | |
| Band Stacking, RGB & False Color Images, and Interactive Widgets in Python | Bridget | |
| 15:00 | BREAK | |
| Plot Spectral Signatures | Bridget | |
| Calculate NDVI | Bridget | |
| Calculate Other Indices; Small Group Coding | Megan | |
| 17:30 | GitHub Workflow | Naupaka Zimmerman |
| 18:00 | End of Day Wrap Up | Megan |
Additional Information
This morning, we will be touring the NEON facilities including several labs. Please wear long pants and close-toed shoes to conform to lab safety standards. Many individuals find the temperature of the classroom where the Data Institute is held to be cooler than they prefer. We recommend you bring a sweater or light jacket with you. You will have the opportunity to eat your lunch on an outdoor patio - hats, sunscreen, and sunglasses may be appreciated.
Additional Resources
Participants looking for more background on the HDF5 format may find these tutorials useful.
- Hierarchical Data Formats - What is HDF5? tutorial
- Explore HDF5 Files using the HDF5View Tool tutorial
During the 2016 Data Institute, Dr. Dave Schimel gave a presentation on the importance of "Big Data, Open Data, and Biodiversity" and is very much related to the themes of this Data Institute. If interested you can watch the video here.
Tuesday: Lidar Data & Reproducible Workflows
In the morning, we will focus on data workflows, organization and automation as a means to write more efficient, usable code. Later, we will review the basics of discrete return and full waveform lidar data. We will then work with some NEON lidar derived raster data products.
Learning Objectives
After completing these activities, you will be able to:
- Explain the difference between active and passive sensors.
- Explain the difference between discrete return and full waveform LiDAR.
- Describe applications of LiDAR remote sensing data in the natural sciences.
- Describe several NEON LiDAR remote sensing data products.
- Explain why modularization is important and supports efficient coding practices.
- How to modularize code using functions.
- Integrate basic automation into your existing data workflow.
Morning: Reproducible Workflows
| Time | Topic | Instructor/Location |
|---|---|---|
| 8:00 | Automate & Modularize Workflows | Naupaka Zimmerman |
| 10:30 | BREAK | |
| 10:45 | Automate & Modularize Workflows, cont. | |
| 12:00 | LUNCH | Classroom/Patio |
Afternoon: Lidar
| Time | Topic | Instructor/Location |
|---|---|---|
| 13:00 | An Introduction to Discrete Lidar (video) | Tristan Goulden |
| An Introduction to Waveform Lidar (related video) | Keith Krause | |
| OpenTopography as a Data Source (download PDF) | Benjamin Gross | |
| 14:00 | Rasters & TIFF tags | Tristan |
| 14:15 | Classify a Raster using Threshold Values | Bridget |
| Mask a Raster using Threshold Values | Bridget | |
| Create a Hillshade from a Terrain Raster in Python | Bridget | |
| 15:00 | BREAK | |
| 15:15 | Lidar Small Group Coding Activity | Tristan & Bridget |
| 18:00 | End of Day Wrap Up | Megan Jones |
Wednesday: Uncertainty
Today, we will focus on the importance of uncertainty when using remote sensing data.
Learning Objectives
After completing these activities, you will be able to:
- Measure the differences between a metric derived from remote sensing data and the same metric derived from data collected on the ground.
| Time | Topic | Instructor/Location |
|---|---|---|
| 8:00 | Uncertainty & Lidar Data Presentation (video) | Tristan Gouldan |
| 8:40 | Exploring Uncertainty in LiDAR Data | Tristan |
| 10:30 | BREAK | |
| 10:45 | Lidar Uncertainty cont. | Tristan |
| 12:00 | LUNCH | Classroom/Patio |
| 13:00 | Spectral Calibration & Uncertainty Presentation (video) | Nathan Leisso |
| 13:30 | Hyperspectral Variation Uncertainty Analysis in Python | Tristan |
| Assessing Spectrometer Accuracy using Validation Tarps with Python | Tristan | |
| 15:00 | BREAK | |
| 15:50 | Uncertainty in BRDF Flight Data Products at Three Locations presentation | Amanda Roberts |
| Hyperspectral Validation, cont. | Tristan | |
| 18:00 | End of Day Wrap Up | Megan Jones |
Thursday: Applications
On Thursday, we will begin to think about the different types of analysis that we can do by fusing LiDAR and hyperspectral data products.
Learning Objectives
After completing these activities, you will be able to:
- Classify different spectra from a hyperspectral data product
- Map the crown of trees from hyperspectral & lidar data
- Calculate biomass of vegetation
| Time | Topic | Instructor/Location |
|---|---|---|
| 8:00 | Applications of Remote Sensing | Paul Gader |
| 9:00 | NEON Vegetation Data (related video, download PDF ) | Katie Jones |
| NEON Foliar Chemistry & Soil Chemistry Data and Microbial Data (video) | Samantha Weintraub | |
| 9:40 | Classification of Spectra | Paul |
| Classification of Hyperspectral Data with Ordinary Least Squares in Python, (download PDF ) | Paul | |
| Classification of Hyperspectral Data with Principal Components Analysis in Python, (download PDF ) | Paul | |
| Classification of Hyperspectral Data with SciKit & SVM in Python, (download PDF ) | Paul | |
| 10:30 | BREAK | |
| 10:45 | Classification of Spectra, cont. | Paul |
| 12:00 | LUNCH | Classroom/Patio |
| 13:00 | Tree Crown Mapping | Paul |
| 15:00 | BREAK | |
| 15:15 | Biomass Calculations | Tristan Goulden |
| 17:30 | Capstone Brainstorm & Group Selection | Megan Jones |
Calculate Vegetation Biomass from LiDAR Data in Python
In this tutorial, we will calculate the biomass for a section of the SJER site. We will be using the Canopy Height Model discrete LiDAR data product as well as NEON field data on vegetation data. This tutorial will calculate Biomass for individual trees in the forest.
Objectives
After completing this tutorial, you will be able to:
- Learn how to apply a Gaussian smoothing kernel for high-frequency spatial filtering
- Apply a watershed segmentation algorithm for delineating tree crowns
- Calculate biomass predictor variables from a CHM
- Set up training data for biomass predictions
- Apply a Random Forest machine learning model to calculate biomass
Install Python Packages
- gdal
- scipy
- scikit-learn
- scikit-image
The following packages should be part of the standard conda installation:
- os
- sys
- numpy
- matplotlib
Download Data
If you have already downloaded the data set for the Data Institute, you have the data for this tutorial within the SJER directory. If you would like to just download the data for this tutorial use the following links.
Download the Training Data: SJER_Biomass_Training.csv
Download the SJER Canopy Height Model Tile: NEON_D17_SJER_DP3_256000_4106000_CHM.tif
In this tutorial, we will calculate the biomass for a section of the SJER site. We will be using the Canopy Height Model discrete LiDAR data product as well as NEON field data on vegetation data. This tutorial will calculate biomass for individual trees in the forest.
The calculation of biomass consists of four primary steps:
- Delineate individual tree crowns
- Calculate predictor variables for all individual trees
- Collect training data
- Apply a Random Forest regression model to estimate biomass from the predictor variables
In this tutorial we will use a watershed segmentation algorithm for delineating tree crowns (step 1) and and a Random Forest (RF) machine learning algorithm for relating the predictor variables to biomass (part 4). The predictor variables were selected following suggestions by Gleason et al. (2012) and biomass estimates were determined from DBH (diameter at breast height) measurements following relationships given in Jenkins et al. (2003).
Get Started
First, we will import some Python packages required to run various parts of the script:
import os, sys
import gdal, osr
import numpy as np
import matplotlib.pyplot as plt
from scipy import ndimage as ndi
%matplotlib inline
Next, we will add libraries from scikit-learn which will help with the watershed delination, determination of predictor variables and random forest algorithm
#Import biomass specific libraries
from skimage.morphology import watershed
from skimage.feature import peak_local_max
from skimage.measure import regionprops
from sklearn.ensemble import RandomForestRegressor
We also need to specify the directory where we will find and save the data needed for this tutorial. You may need to change this line to follow a different working directory structure, or to suit your local machine. I have decided to save my data in the following directory:
data_path = os.path.abspath(os.path.join(os.sep,'neon_biomass_tutorial','data'))
data_path
'D:\\neon_biomass_tutorial\\data'
Define functions
Now we will define a few functions that allow us to more easily work with the NEON data.
-
plot_band_array: function to plot NEON geospatial raster data
#Define a function to plot a raster band
def plot_band_array(band_array,image_extent,title,cmap_title,colormap,colormap_limits):
plt.imshow(band_array,extent=image_extent)
cbar = plt.colorbar(); plt.set_cmap(colormap); plt.clim(colormap_limits)
cbar.set_label(cmap_title,rotation=270,labelpad=20)
plt.title(title); ax = plt.gca()
ax.ticklabel_format(useOffset=False, style='plain')
rotatexlabels = plt.setp(ax.get_xticklabels(),rotation=90)
-
array2raster: function to convert a numpy array to a geotiff file
def array2raster(newRasterfn,rasterOrigin,pixelWidth,pixelHeight,array,epsg):
cols = array.shape[1]
rows = array.shape[0]
originX = rasterOrigin[0]
originY = rasterOrigin[1]
driver = gdal.GetDriverByName('GTiff')
outRaster = driver.Create(newRasterfn, cols, rows, 1, gdal.GDT_Float32)
outRaster.SetGeoTransform((originX, pixelWidth, 0, originY, 0, pixelHeight))
outband = outRaster.GetRasterBand(1)
outband.WriteArray(array)
outRasterSRS = osr.SpatialReference()
outRasterSRS.ImportFromEPSG(epsg)
outRaster.SetProjection(outRasterSRS.ExportToWkt())
outband.FlushCache()
-
raster2array: function to conver rasters to an array
def raster2array(geotif_file):
metadata = {}
dataset = gdal.Open(geotif_file)
metadata['array_rows'] = dataset.RasterYSize
metadata['array_cols'] = dataset.RasterXSize
metadata['bands'] = dataset.RasterCount
metadata['driver'] = dataset.GetDriver().LongName
metadata['projection'] = dataset.GetProjection()
metadata['geotransform'] = dataset.GetGeoTransform()
mapinfo = dataset.GetGeoTransform()
metadata['pixelWidth'] = mapinfo[1]
metadata['pixelHeight'] = mapinfo[5]
metadata['ext_dict'] = {}
metadata['ext_dict']['xMin'] = mapinfo[0]
metadata['ext_dict']['xMax'] = mapinfo[0] + dataset.RasterXSize/mapinfo[1]
metadata['ext_dict']['yMin'] = mapinfo[3] + dataset.RasterYSize/mapinfo[5]
metadata['ext_dict']['yMax'] = mapinfo[3]
metadata['extent'] = (metadata['ext_dict']['xMin'],metadata['ext_dict']['xMax'],
metadata['ext_dict']['yMin'],metadata['ext_dict']['yMax'])
if metadata['bands'] == 1:
raster = dataset.GetRasterBand(1)
metadata['noDataValue'] = raster.GetNoDataValue()
metadata['scaleFactor'] = raster.GetScale()
# band statistics
metadata['bandstats'] = {} # make a nested dictionary to store band stats in same
stats = raster.GetStatistics(True,True)
metadata['bandstats']['min'] = round(stats[0],2)
metadata['bandstats']['max'] = round(stats[1],2)
metadata['bandstats']['mean'] = round(stats[2],2)
metadata['bandstats']['stdev'] = round(stats[3],2)
array = dataset.GetRasterBand(1).ReadAsArray(0,0,
metadata['array_cols'],
metadata['array_rows']).astype(np.float)
array[array==int(metadata['noDataValue'])]=np.nan
array = array/metadata['scaleFactor']
return array, metadata
else:
print('More than one band ... function only set up for single band data')
-
crown_geometric_volume_pct: function to get the tree height and crown volume percentiles
def crown_geometric_volume_pct(tree_data,min_tree_height,pct):
p = np.percentile(tree_data, pct)
tree_data_pct = [v if v < p else p for v in tree_data]
crown_geometric_volume_pct = np.sum(tree_data_pct - min_tree_height)
return crown_geometric_volume_pct, p
-
get_predictors: function to get the predictor variables from the biomass data
def get_predictors(tree,chm_array, labels):
indexes_of_tree = np.asarray(np.where(labels==tree.label)).T
tree_crown_heights = chm_array[indexes_of_tree[:,0],indexes_of_tree[:,1]]
full_crown = np.sum(tree_crown_heights - np.min(tree_crown_heights))
crown50, p50 = crown_geometric_volume_pct(tree_crown_heights,tree.min_intensity,50)
crown60, p60 = crown_geometric_volume_pct(tree_crown_heights,tree.min_intensity,60)
crown70, p70 = crown_geometric_volume_pct(tree_crown_heights,tree.min_intensity,70)
return [tree.label,
np.float(tree.area),
tree.major_axis_length,
tree.max_intensity,
tree.min_intensity,
p50, p60, p70,
full_crown,
crown50, crown60, crown70]
Canopy Height Data
With everything set up, we can now start working with our data by define the file path to our CHM file. Note that you will need to change this and subsequent filepaths according to your local machine.
chm_file = os.path.join(data_path,'NEON_D17_SJER_DP3_256000_4106000_CHM.tif')
chm_file
'D:\\neon_biomass_tutorial\\data\\NEON_D17_SJER_DP3_256000_4106000_CHM.tif'
When we output the results, we will want to include the same file information as the input, so we will gather the file name information.
#Get info from chm file for outputting results
chm_name = os.path.basename(chm_file)
Now we will get the CHM data...
chm_array, chm_array_metadata = raster2array(chm_file)
..., plot it, and save the figure.
#Plot the original CHM
plt.figure(1)
#Plot the CHM figure
plot_band_array(chm_array,chm_array_metadata['extent'],
'Canopy Height Model',
'Canopy Height (m)',
'Greens',[0, 9])
plt.savefig(os.path.join(data_path,chm_name.replace('.tif','.png')),dpi=300,orientation='landscape',
bbox_inches='tight',
pad_inches=0.1)
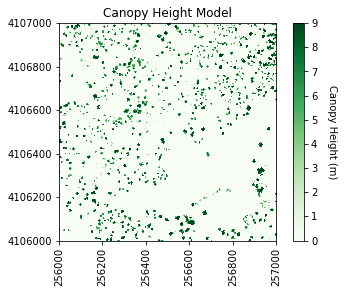
It looks like SJER primarily has low vegetation with scattered taller trees.
Create Filtered CHM
Now we will use a Gaussian smoothing kernal (convolution) across the data set to remove spurious high vegetation points. This will help ensure we are finding the treetops properly before running the watershed segmentation algorithm.
For different forest types it may be necessary to change the input parameters. Information on the function can be found in the SciPy documentation.
Of most importance are the second and fifth inputs. The second input defines the standard deviation of the Gaussian smoothing kernal. Too large a value will apply too much smoothing, too small and some spurious high points may be left behind. The fifth, the truncate value, controls after how many standard deviations the Gaussian kernal will get cut off (since it theoretically goes to infinity).
#Smooth the CHM using a gaussian filter to remove spurious points
chm_array_smooth = ndi.gaussian_filter(chm_array,2,mode='constant',cval=0,truncate=2.0)
chm_array_smooth[chm_array==0] = 0
Now save a copy of filtered CHM. We will later use this in our code, so we'll output it into our data directory.
#Save the smoothed CHM
array2raster(os.path.join(data_path,'chm_filter.tif'),
(chm_array_metadata['ext_dict']['xMin'],chm_array_metadata['ext_dict']['yMax']),
1,-1,np.array(chm_array_smooth,dtype=float),32611)
Determine local maximums
Now we will run an algorithm to determine local maximums within the image. Setting indices to False returns a raster of the maximum points, as opposed to a list of coordinates. The footprint parameter is an area where only a single peak can be found. This should be approximately the size of the smallest tree. Information on more sophisticated methods to define the window can be found in Chen (2006).
#Calculate local maximum points in the smoothed CHM
local_maxi = peak_local_max(chm_array_smooth,indices=False, footprint=np.ones((5, 5)))
Our new object local_maxi is an array of boolean values where each pixel is identified as either being the local maximum (True) or not being the local maximum (False).
local_maxi
array([[False, False, False, ..., False, False, False],
[False, False, False, ..., False, False, False],
[False, False, False, ..., False, False, False],
...,
[False, False, False, ..., False, False, False],
[False, False, False, ..., False, False, False],
[False, False, False, ..., False, False, False]])
This is helpful, but it can be difficult to visualize boolean values using our typical numeric plotting procedures as defined in the plot_band_array function above. Therefore, we will need to convert this boolean array to an numeric format to use this function. Booleans convert easily to integers with values of False=0 and True=1 using the .astype(int) method.
local_maxi.astype(int)
array([[0, 0, 0, ..., 0, 0, 0],
[0, 0, 0, ..., 0, 0, 0],
[0, 0, 0, ..., 0, 0, 0],
...,
[0, 0, 0, ..., 0, 0, 0],
[0, 0, 0, ..., 0, 0, 0],
[0, 0, 0, ..., 0, 0, 0]])
Next we can plot the raster of local maximums by coercing the boolean array into an array of integers inline. The following figure shows the difference in finding local maximums for a filtered vs. non-filtered CHM.
We will save the graphics (.png) in an outputs folder sister to our working directory and data outputs (.tif) to our data directory.
#Plot the local maximums
plt.figure(2)
plot_band_array(local_maxi.astype(int),chm_array_metadata['extent'],
'Maximum',
'Maxi',
'Greys',
[0, 1])
plt.savefig(data_path+chm_name[0:-4]+ '_Maximums.png',
dpi=300,orientation='landscape',
bbox_inches='tight',pad_inches=0.1)
array2raster(data_path+'maximum.tif',
(chm_array_metadata['ext_dict']['xMin'],chm_array_metadata['ext_dict']['yMax']),
1,-1,np.array(local_maxi,dtype=np.float32),32611)
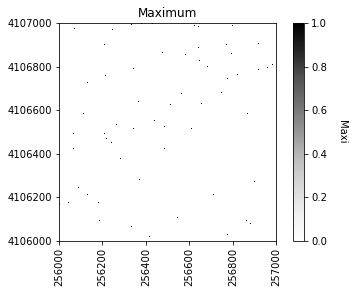
If we were to look at the overlap between the tree crowns and the local maxima from each method, it would appear a bit like this raster.
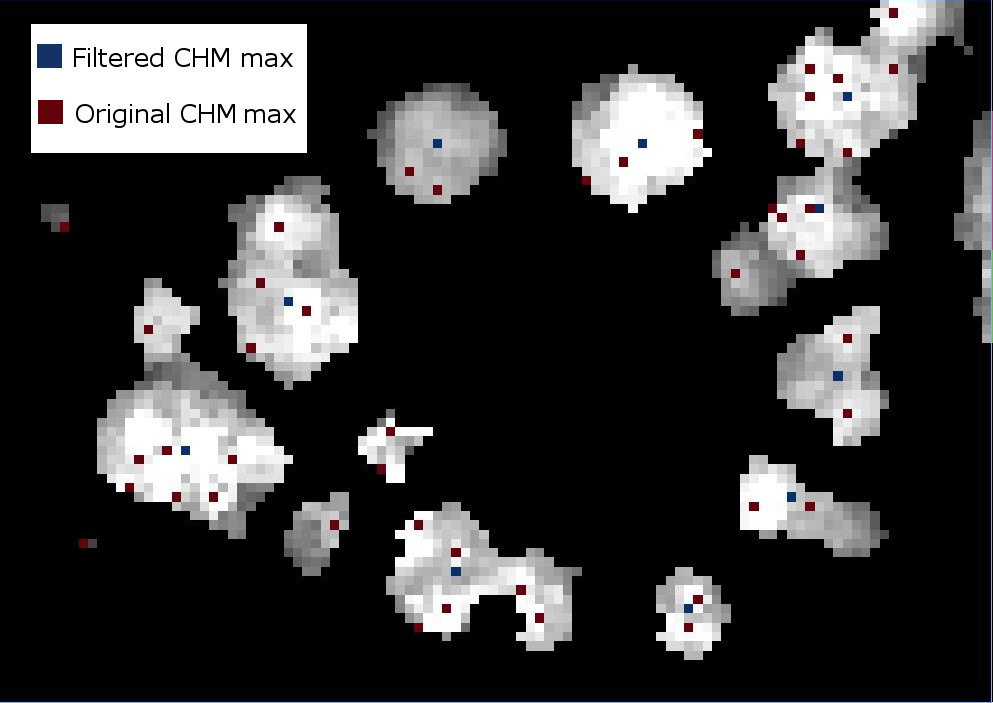
Apply labels to all of the local maximum points
#Identify all the maximum points
markers = ndi.label(local_maxi)[0]
Next we will create a mask layer of all of the vegetation points so that the watershed segmentation will only occur on the trees and not extend into the surrounding ground points. Since 0 represent ground points in the CHM, setting the mask to 1 where the CHM is not zero will define the mask
#Create a CHM mask so the segmentation will only occur on the trees
chm_mask = chm_array_smooth
chm_mask[chm_array_smooth != 0] = 1
Watershed segmentation
As in a river system, a watershed is divided by a ridge that divides areas. Here our watershed are the individual tree canopies and the ridge is the delineation between each one.
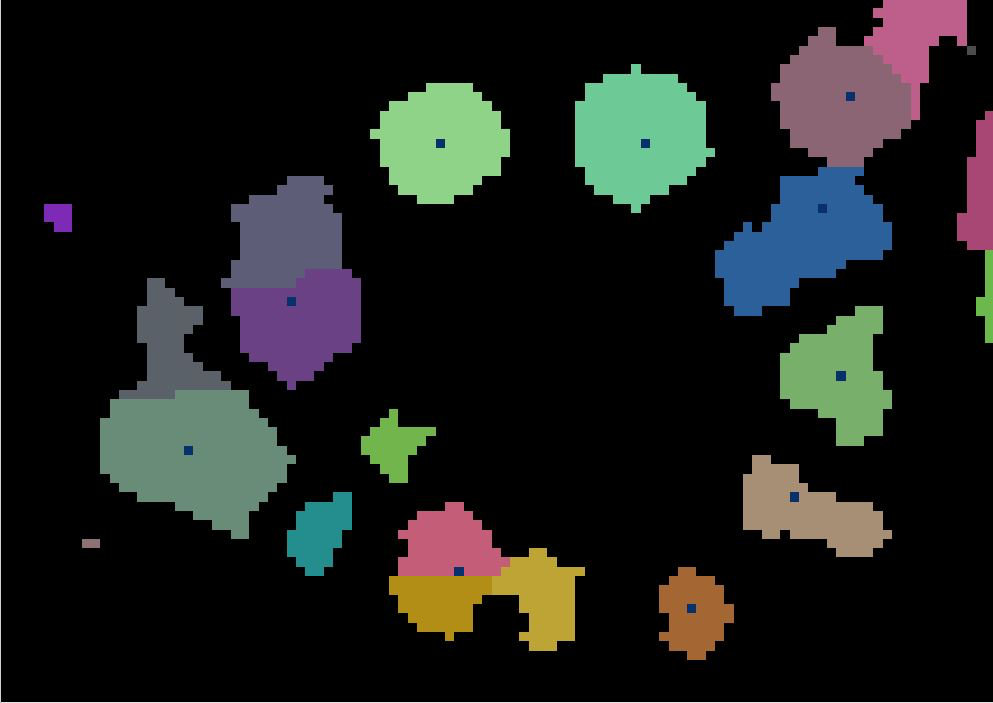
Next, we will perform the watershed segmentation which produces a raster of labels.
#Perform watershed segmentation
labels = watershed(chm_array_smooth, markers, mask=chm_mask)
labels_for_plot = labels.copy()
labels_for_plot = np.array(labels_for_plot,dtype = np.float32)
labels_for_plot[labels_for_plot==0] = np.nan
max_labels = np.max(labels)
#Plot the segments
plot_band_array(labels_for_plot,chm_array_metadata['extent'],
'Crown Segmentation','Tree Crown Number',
'Spectral',[0, max_labels])
plt.savefig(data_path+chm_name[0:-4]+'_Segmentation.png',
dpi=300,orientation='landscape',
bbox_inches='tight',pad_inches=0.1)
array2raster(data_path+'labels.tif',
(chm_array_metadata['ext_dict']['xMin'],
chm_array_metadata['ext_dict']['yMax']),
1,-1,np.array(labels,dtype=float),32611)
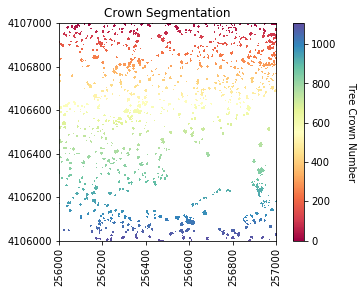
Now we will get several properties of the individual trees will be used as predictor variables.
#Get the properties of each segment
tree_properties = regionprops(labels,chm_array)
Now we will get the predictor variables to match the (soon to be loaded) training data using the get_predictors function defined above. The first column will be segment IDs, the rest will be the predictor variables, namely the tree label, area, major_axis_length, maximum height, minimum height, height percentiles (p50, p60, p70), and crown geometric volume percentiles (full and percentiles 50, 60, and 70).
predictors_chm = np.array([get_predictors(tree, chm_array, labels) for tree in tree_properties])
X = predictors_chm[:,1:]
tree_ids = predictors_chm[:,0]
Training data
We now bring in the training data file which is a simple CSV file with no header. If you haven't yet downloaded this, you can scroll up to the top of the lesson and find the Download Data section. The first column is biomass, and the remaining columns are the same predictor variables defined above. The tree diameter and max height are defined in the NEON vegetation structure data along with the tree DBH. The field validated values are used for training, while the other were determined from the CHM and camera images by manually delineating the tree crowns and pulling out the relevant information from the CHM.
Biomass was calculated from DBH according to the formulas in Jenkins et al. (2003).
#Get the full path + training data file
training_data_file = os.path.join(data_path,'SJER_Biomass_Training.csv')
#Read in the training data csv file into a numpy array
training_data = np.genfromtxt(training_data_file,delimiter=',')
#Grab the biomass (Y) from the first column
biomass = training_data[:,0]
#Grab the biomass predictors from the remaining columns
biomass_predictors = training_data[:,1:12]
Random Forest classifiers
We can then define parameters of the Random Forest classifier and fit the predictor variables from the training data to the Biomass estimates.
#Define parameters for the Random Forest Regressor
max_depth = 30
#Define regressor settings
regr_rf = RandomForestRegressor(max_depth=max_depth, random_state=2)
#Fit the biomass to regressor variables
regr_rf.fit(biomass_predictors,biomass)
RandomForestRegressor(bootstrap=True, ccp_alpha=0.0, criterion='mse',
max_depth=30, max_features='auto', max_leaf_nodes=None,
max_samples=None, min_impurity_decrease=0.0,
min_impurity_split=None, min_samples_leaf=1,
min_samples_split=2, min_weight_fraction_leaf=0.0,
n_estimators=100, n_jobs=None, oob_score=False,
random_state=2, verbose=0, warm_start=False)
We will now apply the Random Forest model to the predictor variables to estimate biomass
#Apply the model to the predictors
estimated_biomass = regr_rf.predict(X)
To output a raster, pre-allocate (copy) an array from the labels raster, then cycle through the segments and assign the biomass estimate to each individual tree segment.
#Set an out raster with the same size as the labels
biomass_map = np.array((labels),dtype=float)
#Assign the appropriate biomass to the labels
biomass_map[biomass_map==0] = np.nan
for tree_id, biomass_of_tree_id in zip(tree_ids, estimated_biomass):
biomass_map[biomass_map == tree_id] = biomass_of_tree_id
Calculate Biomass
Collect some of the biomass statistics and then plot the results and save an output geotiff.
#Get biomass stats for plotting
mean_biomass = np.mean(estimated_biomass)
std_biomass = np.std(estimated_biomass)
min_biomass = np.min(estimated_biomass)
sum_biomass = np.sum(estimated_biomass)
print('Sum of biomass is ',sum_biomass,' kg')
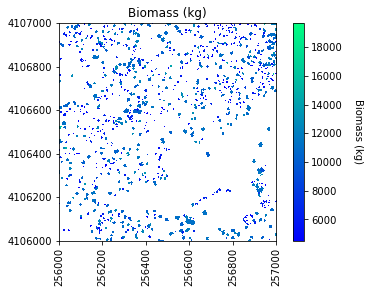
# Plot the biomass!
plt.figure(5)
plot_band_array(biomass_map,chm_array_metadata['extent'],
'Biomass (kg)','Biomass (kg)',
'winter',
[min_biomass+std_biomass, mean_biomass+std_biomass*3])
# Save the biomass figure; use the same name as the original file, but replace CHM with Biomass
plt.savefig(os.path.join(data_path,chm_name.replace('CHM.tif','Biomass.png')),
dpi=300,orientation='landscape',
bbox_inches='tight',
pad_inches=0.1)
# Use the array2raster function to create a geotiff file of the Biomass
array2raster(os.path.join(data_path,chm_name.replace('CHM.tif','Biomass.tif')),
(chm_array_metadata['ext_dict']['xMin'],chm_array_metadata['ext_dict']['yMax']),
1,-1,np.array(biomass_map,dtype=float),32611)
Sum of biomass is 7249752.02745825 kg
Friday: Applications in Remote Sensing
Today, you will use all of the skills you've learned at the Institute to work on a group project that uses NEON or related data!
Learning Objectives
During this activity you will:
- Apply the skills that you have learned to process data using efficient coding practices.
- Apply your understanding of remote sensing data and use it to address a science question of your choice.
- Implement version control and collaborate with your colleagues through the GitHub platform.
| Time | Topic | Location |
|---|---|---|
| 9:00 | Groups begin work on capstone projects | Breakout rooms |
| Instructors available on an as needed basis for consultation & help | ||
| 12:00 | Lunch | Classroom/Patio |
| Groups continue to work on capstone projects | Breakout rooms | |
| 16:30 | End of day wrap up | Classroom |
| 18:00 | Time to leave the building (if group opts to work after wrap up) |
Additional Resources
Saturday: Capstone Projects
| Time | Topic | Instructor |
|---|---|---|
| 9:00 | Presentations Start | |
| 11:30 | Final Questions & Institute Debrief | |
| 12:00 | Lunch | |
| 13:00 | End |